Abstract
NY-ESO-1 is a SEREX-defined cancer-testis antigen of which several MHC I, but only few MHC II–restricted epitopes have been identified. Searching for highly promiscuous MHC II–restricted peptides that might be suitable as a CD4+ stimulating vaccine for many patients, we used the SYFPEITHI algorithm and identified an NY-ESO-1–derived pentadecamer epitope (p134–148) that induced specific CD4+ T-cell responses restricted to the HLA-DRB1 subtypes *0101, *0301, *0401, and *0701 that have a cumulative prevalence of 40% in the Caucasian population. The DR restriction of the p134–148 pentadecamer was demonstrated by inhibition with an HLA-DR antibody and a functional peptide displacement titration assay with the pan-DR-binding T-helper epitope PADRE as the competitor. The natural processing and presentation of this epitope was demonstrated by recognition of an NY-ESO-1+ melanoma cell line by T cells with specificity for p134–148. The pentadecamer p134–148 was able to induce CD4+ responses in 4/38 cancer patients tested. However, no strict correlation was found between CD4+ T-cell responses against p134–148 reactivity and anti-NY-ESO-1 antibody titers in the serum of patients, suggesting that CD4+ and B-cell responses are regulated independently. In conclusion, p134–148 holds promise as a broadly applicable peptide vaccine for patients with NY-ESO-1–positive neoplasms.
Similar content being viewed by others
Introduction
According to their expression pattern and the specificity of the immune responses they evoke, antigens expressed by human neoplasms can be classified into different groups [1]. These include the so-called shared tumor antigens, the differentiation antigens (including the idiotypes of B-cell lymphomas), the products of viral, mutated, differentially spliced, overexpressed and amplified genes, as well as common autoantigens expressed by the malignant cells of a tumor. The most attractive candidates for vaccine development are the so-called shared tumor-specific antigens because they are expressed in a broad spectrum of different human neoplasms. This group includes the CTL-reactive MAGE [2], BAGE [3], and GAGE [4] families, as well as HOM-MEL-40/SSX-2 [5], the other SSX family members [6], NY-ESO-1 [7], HOM-TES-14/SCP-1 [8], CT-7 [9], and HOM-TES-85 [10], all of which have been defined using SEREX, the serological identification of antigens by recombinant expression cloning [11]. It is enigmatic that the expression of all of the so-called shared tumor antigens in humans that have been molecularly defined to date by cellular [12] and serological techniques [13] is restricted to different types of cancers and normal testis. Therefore the term “cancer-testis antigens” (CTAs) [7] or “cancer-germline antigens” [14] has been coined for them and the term “cancer-testis genes” for their encoding genes.
NY-ESO-1 is one of the CTAs with the broadest expression pattern [15], and unlike many other cancer-testis antigens, elicits both humoral and cellular immune responses in a high proportion of patients with NY-ESO-1+ cancers [16]. Because NY-ESO-1 was originally identified by SEREX, its capacity to induce T-cell responses in addition to antibody responses had to be demonstrated a posteriori. Pursuing different strategies of what has been termed “reverse T-cell immunology,” several MHC I–restricted NY-ESO-1 epitopes have been identified, most of them for the HLA-A2 haplotype which is shared by 40% of the Caucasian population.
Increasing evidence from both human and animal studies indicates that CD4+ T cells play a central role in initiating and maintaining host immune responses against cancer [17, 18]. This, together with the fact that in vaccine trials with MHC I–restricted peptides, CD8+ responses were readily generated, but were associated with only limited clinical responses [19], has raised an increased interest for vaccine strategies employing MHC II–restricted peptides either alone or in combination with CD8+ stimulating MHC I–restricted peptides. However, in contrast to the situation with HLA-A2, in the case of MHC I molecules, there is no similarly predominant MHC II haplotype in the Caucasian population. Within the MHC II genes, the DR locus is the one with the highest polymorphism. In spite of the fact that there are only three different alleles coding for the α chain of the DR molecule, genes encoding the β chain have a highly multiple allelomorphy. Consequently, the number of different DR molecules is larger compared with other class II families (data derived from the HLA Informatics Group of the Anthony Nolan Trust updated in October 2002, see www.anthonynolan.org.uk/HIG/). On the other hand, the variability of the individual alleles is limited compared with the MHC class I alleles [20]. Because of this, the number of different HLA-DR–binding epitopes is expected to be smaller than that of MHC I–binding epitopes, and the respective peptides should bind more promiscuously to different allelic subtypes, which is in sharp contrast to class I–restricted epitopes. HLA-DR binding is best studied among the MHC II–restricted epitopes. Nevertheless, only few HLA-DR–restricted NY-ESO-1 epitopes have been defined to date [21, 22, 23, 24, 25, 26]. It is therefore necessary to define new epitopes with the objective of covering a broader range of different HLA-DR subtypes in order to enlarge the number of patients suitable for vaccination with the corresponding peptides.
Aiming at tumor antigen–derived epitopes binding to HLA-DR subtypes that cover a significant proportion of the population, we decided to pursue a strategy that allows for the identification of binding properties which are shared by several HLA-DR haplotypes. This should be feasible, because class II–restricted peptides have a less stringent binding pattern than class I peptides. Employing the SYPEITHI algorithm [27], it became evident that the four DRB1 molecules *0101, *0301, *0401, and *0701, which have a cumulative prevalence of 40% among the Caucasian population [20], share such peptide-binding properties. Screening the entire amino acid sequence of the NY-ESO-1 molecule with the SYFPEITHI algorithm for binding motifs for these DRB1 subtypes, we were able to identify three peptides with motifs suggesting a promiscuous binding and the capacity to trigger MHC II–restricted T-cell responses. Of these, one pentadecamer peptide of the NY-ESO-1 antigen was identified that fulfills the criteria for a widely applicable HLA-DR–restricted peptide vaccine.
Materials and methods
The study had been approved by the local ethics review committee (Ethikkommission der Ärztekammer des Saarlandes) and was done in accordance with the Declaration of Helsinki. Recombinant DNA work was done with the permission and according to the regulations of the local authorities (Regierung des Saarlandes).
Patients
A total of 41 patients were included in this study, of whom 35 had lung cancer, 2 had pleural mesothelioma, and 4 had breast cancer. All patients gave written informed consent. Most lung cancer patients had measurable disease and were undergoing chemotherapy and/or radiotherapy when tested for anti–NY-ESO-1 T-cell reactivity. Only the four breast cancer patients had been only operated on. The patients were investigated for T-cell responses with their HLA-DR subtypes unknown. The HLA-DR typing was performed only in patients who had shown a T-cell reactivity against NY-ESO-1 and in five nonreactive control patients.
Cell lines
The NY-ESO-1–expressing melanoma cell line SK-MEL-37 was kindly provided by Elisabeth Stockert (LICR, New York). SK-MEL-37 is positive for the HLA-DRB1 subtypes *0101 and *0301 and for DRB3*0202. Me 275, which is homozygous for DRB1*1302 and for DRB3*0301, also expresses NY-ESO-1 and was kindly provided by Daniel Speiser (LICR, Lausanne). SK-MEL-37 and Me 275 are positive for NY-ESO-1 both at the mRNA and the protein level as demonstrated by immunocytology [15]. Cell lines were cultured in RPMI 1640 / 10% FCS, 2-mM l-glutamine and 1% penicillin/streptomycin (GIBCO, Invitrogen, Karlsruhe, Germany).
Analysis of NY-ESO-1 mRNA expression by tumors
Fresh tumor biopsy samples were frozen within 15 min after surgical excision. The transcription of NY-ESO-1 mRNA was checked by RT-PCR, using conditions as described by Chen et al. [28].
Prediction of HLA-binding peptides by the SYFPEITHI algorithm
Peptides were derived on the basis of the previously published NY-ESO-1 sequence [7]. The SYFPEITHI algorithm (www.syfpeithi.de) was used for the prediction of NY-ESO-1 peptides binding to the three DR subtypes B1*0101, *0301, and *0401. As a result we chose the following three peptides predicted to show a maximum range of promiscuous binding to the three selected HLA-DRB1 molecules: p91–105 (sequence YLAMPFATPMEAELA), p134–148 (TIRLTAADHRQLQLS), and p158–172 (LLMWITQCFLPVFLA). Except for PADRE, which consists of 13 amino acids [29], all peptides used in this study were 15 amino acid residues long. In addition, a mix of peptides (all 15 amino acids long) derived from the pp65 of the human CMV was used as positive control, predicted to bind to the HLA-DR subtypes of interest: p32, p117, p243, p269, p299, p510, and p524. Peptides were synthesized following the Fmoc/tBu strategy as described [30]. Purity was >90% as assessed by HPLC and mass spectrometry. All peptides were dissolved completely in a mixture of water and DMSO. The concentration for each peptide during pulsing was 2 µg/ml corresponding to a molarity of 1.05 µM for p91–105, 1.01 µM for p134–148, and 0.977 µM for p158–172. The DMSO concentration was kept consistently <1% (v/v) during APC pulsing.
In vitro stimulation of T cells with peptides
PBMCs from patients were isolated by Ficoll-Paque Plus separation (Amersham Pharmacia Biotech, Uppsala, Sweden). Unseparated PBMCs (1×107) were divided into at least three vials and frozen until used in the T-cell assays. The remaining cells were divided into aliquots of 4×106 cells. Each of these aliquots was pulsed with a different peptide at a concentration of 2 µg peptide per milliliter by incubation for 2 h at 37°C in a volume of 500 µl serum-free medium (RPMI 1640 + 2-mM l-glutamine + 1% penicillin/streptomycin). After pulsing, cells were washed once with serum-free medium. Cells were then suspended for cultivation in X-Vivo 15 (BioWhittaker Europe, Verviers, Belgium) at a density of 4×106 cells per 2 ml and divided into two wells (1 ml/well) of a 48-well Nunclone plate (Nunc, Denmark) after addition of 5 ng/ml IL-7 (R&D Systems, Wiesbaden, Germany) and 10% human AB serum (BioWhittaker). PBMCs, which served both as antigen-presenting cells (APCs) and a source for effector cells in this assay, were incubated at 37°C. After 24 h, 20 U/ml IL-2 (R&D Systems) were added. The cells were incubated for 6 days under occasional microscopic control. In case of strong proliferation, cells were split. On day 7, the first ELISpot assay was performed. To this end, 7.5×104 and 3.75×104 sensitized CD4+ T cells, respectively, were harvested from each well with the prestimulated cells and used as effector cells in the ELISpot assay. The remaining cells were restimulated with autologous PBMCs which had been pulsed with the appropriate peptide under the same conditions as described for day 1 at a CD4+/APC ratio of 1:1. On day 14, a second ELISpot was performed, and the remaining cells were once more stimulated as described for days 1 and 7, and a final ELISpot was performed on day 21.
ELISpot assay
At least two ELISpot assays were performed on days 7 and 14, respectively; i.e., the first assay 1 week after the primary stimulation and the second 1 week after a second stimulation on day 7. For the ELISpot assay, 5×104 autologous PBMCs pulsed with peptide (2 µg/ml for 2 h at 37°C) per well were used as APCs; 1.25×104 or 2.5×104 CD4+ T cells per well were used as effectors. Blocking with an anti-pan-MHC II antibody was used as a control for the MHC class II–restriction of T-cell responses. To this end, 1.5×105 (7.5×104 resp.) SK-MEL-37 were used as APCs and were suspended in 190 µl fresh medium; 10 µl anti-pan human MHC II antibody (clone WR18; SEROTEC, Biozol Diagnostica Vertrieb, Eching, Germany) was added and incubated for 30 min at 37°C. Cells were washed, spun gently, and resuspended in 150 µl X-Vivo 15 medium and dispensed into three wells. To prove the HLA-DR restriction of T-cell responses against p134–148, PBMCs used as APCs in the ELISpot assay were treated similar to the cell lines: 1.5×105 PBMCs were suspended in 50 µl fresh medium, 20 µl anti-HLA-DR antibody (clone L243; Becton Dickinson, Heidelberg, Germany) was added and incubated for 30 min at 37°C. As an additional control, 20 µl of an anti-HLA-DP antibody (clone B7/21; Becton Dickinson) was used. After staining, irrespective of the antibody, the volume was increased to 500 µl X-Vivo 15, and the respective peptide was added at a concentration of 2 µg/ml. For pulsing, cells were incubated for another 2 h at 37°C. Thereafter, cells were washed, spun down gently, and resuspended in 150 µl X-Vivo 15 medium and dispensed into three wells. PBMCs to be used as APCs for the ELISpot assay were irradiated with 30 Gy. APCs and effector cells (each suspended in 50 µl/well) were coincubated for 14–16 h at 37°C in wells that had been precoated the day before (16 h at 4°C) with anti–IFN-γ capture antibody. This antibody as well as the biotinylated anti–IFN-γ used for developing the ELISpot assay was purchased from Mabtech (Mabtech, Nacka, Sweden). The assay procedure followed the supplier’s instructions with an additional blocking step with 10% (v/v) human AB serum for 1 h at 37°C after the wells had been coated. To enhance the sensitivity of the IFN-γ detection, incubation for 1 h with alkaline phosphatase–conjugated streptavidin (Roche Diagnostics, Mannheim, Germany), diluted 1:2,000 in PBS, followed after unbound anti–IFN-γ biotinylated antibody was washed away. For staining of the IFN-γ spots we used the AP Conjugate Substrate Kit (BIO-RAD Laboratories, Hercules, CA) following the supplier’s instructions. Assays were performed in nitrocellulose-lined 96-well microplates (MAHA S45; Millipore, Bedford, MA). Spots were counted using Bioreader 2000 (BIOSYS, Karben, Germany). All tests were run in triplicate to determine mean values and standard deviation. For ELISpot assays with cell lines, the melanoma cell lines SK-MEL-37 and Me 275 were used as APCs after irradiation with 120 Gy.
Functional peptide displacement assay
T cells were stimulated with NY-ESO-1 p134–148 as described before. To prove the DR-affinity of the T-cell response, a functional peptide displacement assay was used. To this end, the cells to be used as APCs in the ELISpot assay were pulsed with peptide p134–148 which was used for the primary stimulation of the T cells in the presence of increasing concentrations of PADRE, the pan-DR-binding T-helper epitope [29]. For the displacement assay, pulsing of PBMCs was performed by incubation with peptide p134–148 at a concentration of 2 µg/ml, with 2 µg/ml p134–148 in combination with 1 µg/ml PADRE, with 2 µg/ml of both the stimulating and the competing peptide, and with 2 µg/ml of PADRE only. Apart from this pre-ELISpot pulsing of PBMCs, the functional peptide displacement assay was performed and evaluated as described for the ELISpot assay above.
Detection of serum antibodies against NY-ESO-1
Serum antibodies against NY-ESO-1 were assessed semiquantitatively by Western blot as well as by ELISA using Escherichia coli–expressed protein as antigen, as described previously [31].
Immunohistology
Sections from tumor biopsy samples were formalin-fixed immediately after surgical intervention and embedded in paraffin, cut and mounted on slides. Subsequently, the sections were deparaffinized, rehydrated, exposed to hydrogen peroxide (3% v/v) and subjected to antigen retrieval by incubation in 110-mM citrate buffer (pH 6.0) and heating in the microwave oven at 750 VA followed by 350 VA for 5 min. Slides were blocked with a PBS / 1% BSA (v/v) and 20% rabbit serum (v/v) for 20 min at RT and incubated for 30 min at 37°C in monoclonal mouse antihuman HLA-DR / α chain (DAKO, Glostrup, Denmark) diluted 1:50 in PBS / 1% rabbit serum (v/v). The slides were then washed twice in PBS. Then the sections were incubated for 15 min at 37°C in biotinylated rabbit antimouse immunoglobulin (DAKO, Glostrup, Denmark) diluted 1:200 in PBS / 1% rabbit serum (v/v). The slides were washed twice in PBS, incubated for 15 min at 37°C in streptavidin/HRP (DAKO, Glostrup, Denmark) diluted 1:300 in PBS. After a final washing in PBS, DAB substrate (Sigma, St Louis, MO, USA) was added according to the manufacturer’s instructions. Finally, the slides were counterstained with Harris hematoxylin. Negative controls were performed using an irrelevant isotype–matched primary antibody (clone MOPC, murine IgG1-κ; Sigma, St Louis, MO, USA).
Flow cytometry
To characterize effector cells and APCs as well as cell lines, approximately 1×105 cells were harvested on day 0 and 1 day before each ELISpot assay. Cells were washed, spun down gently, and the pellet was resuspended in 90 µl staining buffer. Five microliters of the corresponding antibody (anti-HLA-DR/PE [clone L243], anti-hCD4/FITC, anti-hCD4/PerCP [both from clone SK3], anti-hCD8/PE [clone SK1], anti-hCD45RA [clone L48,]; all obtained from Becton Dickinson, Heidelberg, Germany) and anti-hCCR7/PE (clone #150503; obtained form R&D Systems, Wiesbaden, Germany) were added and mixed, and this suspension was incubated for 20 min at 4°C. After another washing step, the cells were resuspended in a final volume of 300µl CellWash solution (Becton Dickinson). Cells (5×104) were analyzed using a FACScan 80214 (Becton Dickinson).
Results
Identification of a promiscuous HLA-DR epitope
With the goal of developing an NY-ESO-1–derived MHC II peptide vaccine for a broad spectrum of patients we aimed at identifying a peptide with a promiscuous binding pattern for different HLA-DRB1 subtypes. We used the SYFPEITHI algorithm to identify NY-ESO-1 peptides with a high binding affinity score for HLA-DRB1 molecules that have a high prevalence in the Caucasian population. This is the case with the DRB1 subtypes *0101, *0301, and *0401, which occur in 27% of the Caucasian population. Since DRB1*0701, which is found in additional 12% of the population, and DRB1*0401 share most of their anchor positions, we analyzed the affinity of the identified peptides to this subtype, too, even though DRB1*0701 was not yet considered in the SYFPEITHI algorithm when we started our experiments.
SYFPEITHI predicted the following three peptides derived from NY-ESO-1 peptide with a high binding probability: p91–105, p134–148 and p158–172. These peptides were synthesized and used for the stimulation of PBMCs from lung and breast cancer patients, respectively. Whenever enough PBMCs were available, they were tested against all three peptides. However, due to the limited numbers of PBMCs from some patients, only 29 patients were stimulated with p91–105, 21 patients with p158–172, and 38 patients with p134–148.
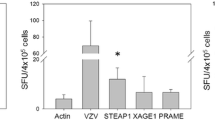
No reproducible T-cell responses were observed after stimulation with the NY-ESO-1 peptides p91–105 and p158–172. In contrast, peptide p134–148 proved to be strongly immunostimulatory. The T cells from 4/38 (11%) patients tested (3/34 with lung cancer and 1/4 with breast cancer) showed a strong response already on day 7 after the first stimulation with p134–148 peptide–loaded autologous PBMCs (Fig. 1A). In addition to these four patients, three patients with lung cancer showed a weak response only after two stimulations (14 days) with spot numbers in the ELISpots being only slightly (but reproducibly and significantly) above the background (data not shown).
Demonstration of HLA-DR restriction
Definitive evidence for the DR restriction of the T-cell response was obtained by blocking the response with an HLA-DR antibody. The lack of toxicity of the antibody was demonstrated by the fact that a MHC I (HLA-A2)–restricted anti-CMV CD8+ T-cell response remained unaffected by this antibody (data not shown). Restriction by HLA-DP was excluded by demonstrating that the anti-HLA-DP antibodies did not interfere with this response (Fig. 2). In addition, a functional peptide displacement titration assay was performed. In this assay, a strictly proportional decrease of the number of IFN-γ–releasing T cells after stimulation with p134–148 was observed in the ELISpot assay when the APCs were pulsed with increasing concentrations of the pan-DR-binding peptide (PADRE) and constant concentrations of the p134–148 peptide. The capacity of PADRE to displace the NY-ESO-1–derived peptide 134–148, with which it competes for binding to the HLA-DR molecules, is shown in Fig. 3. In a reverse competition assay, p134–148 was also capable of displacing PADRE: a 50% reduction of IFN-γ spots in the ELISpot assay of PADRE-stimulated T cells was achieved with APCs pulsed with 2 μg/ml p134–148, corresponding to an IC50 of p134–148 of 1.01 μM (data not shown).
HLA-DR restriction of the T-cell response induced by p134–148. Preincubation of the stimulator cells (autologous PBMCs) with the anti–HLA-DR antibody L243 resulted in blocking of the p134–148–induced T-cell response, while preincubation of the stimulator cells with the anti–HLA-DP antibody B7/21 had no effect
Functional peptide displacement assay. Pulsing of the ELISpot APCs with peptide mixtures containing increasing percentages of the pan-DR-binding T-helper epitope PADRE and reciprocally decreasing percentages of p134–148 molecules resulted in reduced numbers of spots generated by IFN-γ–secreting CD4+ T cells that had been prestimulated with NY-ESO-1–derived p134–148. Solid diamond after 7 days, solid square after 14 days
Determination of HLA-DRB1 subtypes recognized by reactive T cells
To further delineate the HLA-DR restriction of the NY-ESO-1–derived peptide p134–148, the HLA-DR subtype of patients responding to this peptide and of some nonresponding patients who served as controls were determined by HLA-SSP PCR. As can be seen from Table 1, the responding patients had the HLA-DRB1 subtypes *0101, *0301, *0401, and *0701, confirming the predictions of the SYFPEITHI algorithm concerning HLA-DRB1*0101, *0301, and *0401, and our assumptions made with respect to HLA-DRB1*0701, respectively. That p134–148 is indeed presented by B1*0301 was demonstrated by the fact that p134–148–stimulated T cells from patient BC212 recognized the cell line SK-MEL-37, with which they share only the B1*0301 subtype (Fig. 4). Five of the nonresponding patients were also HLA-DR typed. Three of them had no subtype with a high p134–148 binding score. However, of the remaining two patients, one was typed DRB1*0701/*1501, DRB4*0101, and DRB5*0101, and the other DRB1*0301/*1501, DRB3*0202, and DRB5*0101. The reasons for the failure to detect a DRB1*0701 and *0301–restricted T-cell response to p134–148 in these patients will be discussed later.
Demonstration of the DRB1*0301 restriction of the T-cell response against NY-ESO-1 p134–148. After stimulation with p134–148, T cells from patient BC212 recognized the NY-ESO-1+ cell line SK-MEL-37, but not the NY-ESO-1+ cell line Me 275. The restriction of the T-cell response by DRB1*0301 was demonstrated by the fact that DRB1*0301 is the only HLA-DR subtype shared by both the patient and the cell line SK-MEL-37
Response to NY-ESO-1–expressing cell lines
Having demonstrated an HLA-DR–restricted T-cell response against APCs pulsed with the NY-ESO-1–derived peptide p134–148, it was of interest to prove that this peptide is also processed and presented naturally in an HLA-DR–restricted fashion. To this end, two HLA-DR+ melanoma tumor cell lines expressing the NY-ESO-1 antigen at a similar level (data not shown) were chosen. Flow cytometric analysis of SK-MEL-37 and Me 275 cells stained with antihuman pan-HLA-DR antibody revealed that these cells express HLA-DR, irrespective of the temperature at which antibody binding is performed. Similarly, irradiation with 120 Gy did not influence DR expression. SK-MEL-37 expresses HLA-DRB1*0101/*0301 and DRB3*0202, while Me 275 is homozygous for HLA-DRB1*1302 and DRB3*0301. T cells from a DRB1*0301-positive patient who had been prestimulated with p134–148 specifically recognized SK-MEL-37, but not Me 275, demonstrating the processing and DRB1*0301–restricted presentation of p134–148 by SK-MEL-37 (Fig. 5). An additional pulsing of both cell lines with p134–148 resulted in a doubling of the IFN-γ spots compared with the untreated SK-MEL-37, whereas the Me 275 remained unrecognized (Fig. 5). This provides further evidence for the binding of p134–148 to the DRB1*0301 molecules on the SK-MEL-37 cells, but not to the DRB1*1302 molecules on the Me 275 cells.
Natural processing and presentation of NY-ESO-1 epitope p134–148 by NY-ESO-1+ tumor cells. Cells of the SK-MEL-37 melanoma cell line (heterozygous for HLA-DRB1*0301) were recognized by HLA-DRB1*0301–restricted T cells that had been prestimulated with p134–148. In contrast, Me 275 (homozygous for DRB1*1302) was not recognized. Additional exogenous loading of p134–148 resulted in an augmented T-cell response against SK-MEL-37, but not against Me 275
Characterization of T cells after stimulation with p134–148
Because we did not work with T-cell clones, the characterization of T cells could not distinguish between p134–148 responding and nonresponding T cells, but applied to the bulk of T cells after stimulation with the peptide. A T-cell response to p134–148 did not only induce IFN-γ production, but also resulted in the proliferation and expansion of T cells. This expansion was mainly due to CD4+ T cells as demonstrated by their relative increase within the lymphocyte gate after stimulation with the p134–148 peptide, in particular in patients with a strong peptide-induced T-cell response (Fig. 6). The proliferation and expansion of CD4+ T cells was not observed in nonresponding patients (data not shown). With respect to the state of activation, CD4+ T cells acquired HLA-DR expression and shifted from the naïve state to the central memory state according to the schema suggested by Lanzavecchia [32], while the proportion of effector memory cells remained constant. This behavior was different from that observed after stimulation with a mix of viral MHC II–binding peptides derived from the pp65 antigen of human CMV. The stimulation with these viral peptides resulted in a shift of the CD4+ T cells to the effector memory state (Fig. 6).
Phenotypic characterization of CD4+ cells after stimulation with p134–148. A Stimulation with p134–148 resulted in an increase of CD4+ T cells. B Columns 1 & 3 show the state of activation of CD4+ T cells according to the schema suggested by Lanzavecchia [32] before stimulation. While stimulation with NY-ESO-1 p134–148 (column 2) resulted in shift from the naïve state (open column) toward the central-memory state (black column), stimulation with pp65 derived from CMV (column 4) induced a shift toward the effector-memory state (striped column)
Correlation of HLA-DR–restricted T cells and humoral anti-NY-ESO-1 antibody responses
Of the 38 patients tested for a T-cell response to the NY-ESO-1 epitope p134–148, only 5 (13%) had anti-NY-ESO-1 antibodies in their serum as determined by Western blot and ELISA. Three of these five patients were antibody-positive, but no T-cell response to p134–148 was observed. On the other hand, 2/4 patients (50%) with a p134–148–reactive T-cell response were positive for anti-NY-ESO-1 antibodies in their serum (Table 2).
Discussion
The screening of a large number of patients for CD4+ reactivity against peptides derived from the cancer-associated NY-ESO-1 antigen in this study allows for a solid estimation on the expected frequency of the T-cell responses against the respective antigen among patients with cancer; this is not possible if only two or three patients and/or normal controls are tested, as has been the case in previous studies [21, 33, 34]. Moreover, it enabled us to check for obvious associations of the observed T-cell responses with clinical parameters such as histological origin of the tumor, the tumor stage of the patient, or the type of therapy the patients have received. Furthermore, by including lung and breast cancer patients in this study, we gained insight into the T-cell responses among patients suffering from the human neoplasms with the highest incidence in males and females, respectively.
Investigating 41 patients for CD4+ reactivity against NY-ESO-1, it was not possible to determine the HLA-DRB1 subtype from all patients before starting the stimulation or to create a cell line derived from each patient’s tumor biopsy sample. Thus, we used autologous PBMCs as APCs to assess T-cell reactivity. Due to the limited amount of blood we received from the patients, we decided to use only the IFN-γ ELISpot for the evaluation of the patients’ T-cell response. Different strategies can be pursued for the identification of CD4+ T-cell–stimulating peptides derived from a tumor-associated antigen. One such strategy, employed by Jäger et al. [21] is to synthesize and test overlapping peptides that cover the whole antigenic protein. This approach, if applied to the analysis of the T-cell response of patients, is hindered by the enormous amounts of cells necessary from a given patient to test the entire battery of peptides. To narrow down the spectrum of candidate peptides, screening of HLA-DR transgenic mice might be helpful [24]; however the respective transgenic mice are not commonly available. In this study we followed a different strategy that has already been used successfully by others [35, 36] to narrow down the number of NY-ESO-1–derived peptides with putative DR-reactivity: we used the SYFPEITHI algorithm for an in silico screening of the entire NY-ESO-1 protein for peptides with HLA-DR peptide motifs. Making use of the anchor positions shared by the HLA-DRB1 subtypes *0101, *0301, and *0401, we searched for NY-ESO-1 peptides with binding motifs suggesting a promiscuous binding to these three subtypes. Of the three peptides predicted to fulfill this prerequisite with a high binding score in the SYPEITHI algorithm, two peptides (p91–105 and p158–172) failed to induce reproducible T-cell responses. Several reasons might be responsible for this failure: the binding scores for the peptides p91–105 and p158–172 are in general inferior compared with the epitope p134–148, and/or natural processing might not occur.
In contrast to p91–105 and p158–172, the NY-ESO-1–derived epitope p134–148 induced strong T-cell responses in 4/38 (11%) patients tested. The analysis of the HLA typing of the four responding patients revealed that three of them expressed (heterozygously) the DRB1 subtypes *0101, *0301, or *0401 that had been expected by the SYFPEITHI algorithm. However, one responder was typed positive for HLA-DRB1*0701. Thus, the peptide might mediate a promiscuous binding to *0101, *0301, *0401, and *0701. An important consequence of the putative binding shared by *0701 is that the proportion of individuals expressing at least one of these four HLA-DRB1 alleles amounts to 40% of Caucasians . Including the HLA-DR subtypes of two of the additional three patients with lung cancer, who had shown a weak response only after two stimulations (14 days) with spot numbers in the ELISpots being only slightly (but reproducibly and significantly) above the background (data not shown) and for whom HLA-DR subtyping was possible, would not have broadened the spectrum of p134–148–binding HLA-DR subtypes further, because these two patients shared HLA-DR subtypes with the four patients that had a strong T-cell response (data not shown).
That the observed T-cell responses were mediated by other DRB1 subtypes expressed by the responding patients can not be excluded; similarly, since DRB1*04 individuals coexpress both DRB1 and DRB4 gene products, and DRB1*03 individuals coexpress DRB1 and DRB3 gene products, we can not exclude a restriction by the latter gene products either; in any case this would not diminish the broad range of HLA-DR subtypes binding to p134–148.
Several peptides mostly derived from xenogeneic antigens that are capable of broadly binding to different class II molecules have been reported [29, 34, 37, 38, 39], and limited promiscuity has been reported for cancer-associated antigens such as HER-2/neu [39] and MAGE-A3 [40].
The NY-ESO-1–derived peptide p134–148 with its promiscuous binding to at least four different DRB1 subtypes is partially overlapping with the p139–156 epitope, which has previously been shown to bind to DRB4*0101 / *0103 (DR53) [21]. While there is little data available concerning the characteristic binding patterns of DRB4, it would be interesting to investigate whether the promiscuous binding to HLA-DRB1*0101, *0301, *0401, and *0701 extends beyond B1 and includes also B3, B4 (at least its *0101 subtype), and DRB5. The same might apply to other subtypes that are predicted to bind with high affinity to other NY-ESO-1–derived peptides [23]. The definitive identification of the entire spectrum of p134–148–binding HLA-DR molecules would necessitate the demonstration of the binding of the peptide to isolated HLA-DR molecules, presentation of the peptide by APCs transfected with only one HLA-DR subtype or COS cells transfected with NY-ESO-1 and the HLA-DR gene of interest. Alternatively, the DR subtype can be narrowed down by using APCs that share only one HLA-DR subtype with the effector T cells. In this study, this was the case with DRB1*0301 which was the only subtype shared by the cell line SK-MEL-37 and patient BC212 (Fig. 4).
Zarour et al. [41] defined a large epitope-spanning p119–143 that induces T-cell responses in different DR subtypes. Dividing this epitope into smaller peptides, the authors tried to define the stretch responsible for the induction of the T-cell reaction. Interestingly, no response was observed against p129–143, respectively, and only a weak response to p119–133 and p123–137. From these observations they concluded that all of the 25 amino acids of p119–143 are necessary for a strong T-cell response. In a second study, Zarour et al. [23] demonstrated that the CD4+ T-cell response against this peptide was restricted by the HLA-DRB1 subtypes *0101 and *0701, but not by HLA-DRB1*0301 which is expressed by nearly 10% of the Caucasian population.
Not all individuals tested in this study and expressing the HLA-DRB1 subtypes in question showed a T-cell response to this epitope in vitro. Individual differences in the T-cell reactions of patients against epitopes from tumor antigens are well known: possible reasons for this might be a suboptimal presentation of the peptide by the APCs of the respective patients used in this study. We had to use PBMCs and not DCs as APCs, because only limited numbers of PBMCs from a single patient were available for this study, resulting not only in a smaller number, but also a considerable patient-to-patient variability with regard to the quantity and quality of the APCs in the individual PBMC preparation. Indeed, the percentage of HLA-DR–expressing cells among the PBMCs, which can be regarded as a rough correlate for the number of APCs, varied widely from patient to patient analyzed in this study (5% to 35%; data not shown). Alternatively, the low number of spots observed in three patients (whom we did not count as responders) may also be due to the low peptide concentrations (2 μg/ml) that we used compared with other authors [21, 24, 42] for pulsing APCs in order to avoid nonspecific binding of the peptides.
The HLA-DR restriction of the observed T-cell responses was further supported by the results of a functional displacement assay using the pan-DR-binding T-helper epitope PADRE. There was a significant negative linear correlation (R 2=0.96 and p=0.02 at d7, and R 2=0.97 and p=0.01 at d14; Fig. 3) between the PADRE concentration used during APC pulsing for the ELISpot assay and the number of IFN-γ–secreting T-cells prestimulated with the NY-ESO-1 epitope p134–148. Similar to the competitive binding assay previously described for MHC I–binding peptides [40], this strategy represents a powerful method for the characterization of MHC II–binding peptides. In contrast to approaches which use immunopurified HLA-DR molecules coated onto plates, the displacement of competing epitopes from the surface of cells that are used as APCs represents a more physiological way of inhibition. Moreover, it directly provides functional data and allows for the estimation of the affinity of different MHC II–binding peptides using PADRE as the comparator. However, since binding of PADRE to HLA-DP and HLA-DQ can not be excluded, the definitive demonstration of an HLA-DR restriction of a T-cell response that is inhibited by PADRE must be made by blocking with an HLA-DR antibody. Finally, as discussed above, only the demonstration of the binding of a peptide under study to purified MHC II molecules of a given subtype allows for the definitive determination of a restriction.
Since DRB1*04 individuals coexpress both DRB1 and DRB4 gene products, and DRB1*03 individuals coexpress DRB1 and DRB3 gene products, restriction by the latter gene products can not definitely be excluded. However, the latter is very unlikely because all the coexpressed DRB3, DRB4, and DRB5 gene products are expressed at a much lower level than DRB1 on APCs and (in contrast to DRB1) are not expressed in all individuals. Finally, that p134–148 is indeed presented by B1*0301 was proven by demonstrating that p134–148–stimulated T cells from patient BC212 recognized the cell line SK-MEL-37, with which they share only the subtype B1*0301 (Fig. 4).
Our results obtained with the SK-MEL-37 melanoma cell line suggest that p134–148 is naturally processed and presented on the surface of these cells in the context of the respective MHC II molecules. However, even if tumor cells lack MHC II expression, which is the rule rather than the exception both in lung cancers [43] and breast cancers [44], antitumor immunity may be promoted by cross-priming or cross-presentation of both CD8 and CD4 epitopes of tumor antigens by professional APCs, including DCs [45, 46], which may acquire tumor antigens from necrotic or apoptotic bodies in the tumor microenvironment [47]. This mechanism of immune-directed recognition of tumor would allow specific antitumor CD4+ responses to be beneficial to patients, irrespective of the MHC II status of their tumors.
Of the four responders, two had a high anti-NY-ESO-1 antibody titer. Our observation of T-cell responses in anti-NY-ESO-1–negative patients is in contrast to observations reported by others [21] who observed T-cell responses only in anti-NY-ESO-1 antibody–positive patients. However, the constellation of inducible CD4+ responses in antibody-negative patients might have been missed in those studies, because only two and three patients, respectively, were analyzed.
In summary, using the SYFPEITHI algorithm we succeeded in identifying an NY-ESO-1–derived peptide that can be used for vaccine development aimed at the stimulation of CD4+ cells of patients with a broad spectrum of HLA-DR haplotypes. We expect that the approach pursued in this study will also be successful for other SEREX-defined cancer-testis antigens such as HOM-MEL-40 SSX-2 [5] and HOM-TES-14/SCP-1 [8]. Whether the use of such peptides either alone or as part of a polyvalent peptide vaccine cocktail consisting also of appropriate MHC I–restricted epitopes will result in better clinical responses of the vaccinated patients, can now be tested in clinical studies open to patients with a broad spectrum of HLA-DR subtypes.
Abbreviations
- °C:
-
degree Celsius
- CD:
-
cluster of differentiation
- CTA:
-
cancer-testis antigen
- DC:
-
dendritic cells
- Gy:
-
gray
- mM:
-
millimolar
- ng:
-
nanogram
- PADRE:
-
pan-DR-binding T-helper epitope
- pp65:
-
phosphoprotein 65
- SSP-PCR:
-
sequence-specific primer PCR
- v/v:
-
volume per volume
References
Preuss KD, Zwick C, Bormann C, Neumann F, Pfreundschuh M (2002) Analysis of the B-cell repertoire against antigens expressed by human neoplasms. Immunol Rev 188:43
van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, van den Eynde B, Knuth A, Boon T (1991) A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254:1643
Boel P, Wildmann C, Sensi ML, Brasseur R, Renauld JC, Coulie P, Boon T, van der Bruggen P (1995) BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity 2:167
van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T (1995) A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med 182:689
Tureci O, Sahin U, Schobert I, Koslowski M, Scmitt H, Schild HJ, Stenner F, Seitz G, Rammensee HG, Pfreundschuh M (1996) The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res 56:4766
Gure AO, Wei IJ, Old LJ, Chen YT (2002) The SSX gene family: characterization of 9 complete genes. Int J Cancer 101:448
Chen YT, Scanlan MJ, Sahin U, Tureci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ (1997) A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci U S A 94:1914
Tureci O, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M (1998) Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc Natl Acad Sci U S A 95:5211
Chen YT, Gure AO, Tsang S, Stockert E, Jager E, Knuth A, Old LJ (1998) Identification of multiple cancer/testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci U S A 95:6919
Tureci O, Sahin U, Koslowski M, Buss B, Bell C, Ballweber P, Zwick C, Eberle T, Zuber M, Villena-Heinsen C, Seitz G, Pfreundschuh M (2002) A novel tumour associated leucine zipper protein targeting to sites of gene transcription and splicing. Oncogene 21:3879
Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M (1995) Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A 92:11810
van den Eynde B, van der Bruggen P (1997) T cell defined tumor antigens. Curr Opin Immunol 9:684
Tureci O, Sahin U, Pfreundschuh M (1997) Serological analysis of human tumor antigens: molecular definition and implications. Mol Med Today 3:342
Schultz ES, Lethe B, Cambiaso CL, Van Snick J, Chaux P, Corthals J, Heirman C, Thielemans K, Boon T, van der Bruggen P (2000) A MAGE-A3 peptide presented by HLA-DP4 is recognized on tumor cells by CD4+ cytolytic T lymphocytes. Cancer Res 60:6272
Jungbluth AA, Chen YT, Stockert E, Busam KJ, Kolb D, Iversen K, Coplan K, Williamson B, Altorki N, Old LJ (2001) Immunohistochemical analysis of NY-ESO-1 antigen expression in normal and malignant human tissues. Int J Cancer 92:856
Jager E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A (1998) Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med 187:265
Wang RF, Rosenberg SA (1999) Human tumor antigens for cancer vaccine development. Immunol Rev 170:85
Pardoll DM, Topalian SL (1998) The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol 10:588
Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, Wunderlich JR, Parkhurst MR, Kawakami Y, Seipp CA, Einhorn JH, White DE (1998) Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med 4:321
Albert E (1997) Immungenetik. In: Immunology. Gemsa D, Kalden JR, Resch, K (eds.) Thieme, Heidelberg / New York, p 87
Jäger E, Jager D, Karbach J, Chen YT, Ritter G, Nagata Y, Gnjatic S, Stockert E, Arand M, Old LJ, Knuth A (2000) Identification of NY-ESO-1 epitopes presented by human histocompatibility antigen (HLA)-DRB4*0101–0103 and recognized by CD4(+) T lymphocytes of patients with NY-ESO-1-expressing melanoma. J Exp Med 191:625
Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM (2000) NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res 60:4946
Zarour HM, Maillere B, Brusic V, Coval K, Williams E, Pouvelle-Moratille S, Castelli F, Land S, Bennouna J, Logan T, Kirkwood JM (2002) NY-ESO-1 119–143 is a promiscuous major histocompatibility complex class II T-helper epitope recognized by Th1- and Th2-type tumor-reactive CD4+ T cells. Cancer Res 62:213
Zeng G, Touloukian CE, Wang X, Restifo NP, Rosenberg SA, Wang RF (2000) Identification of CD4+ T cell epitopes from NY-ESO-1 presented by HLA-DR molecules. J Immunol 165:1153
Zeng G, Wang X, Robbins PF, Rosenberg SA, Wang RF (2001) CD4(+) T cell recognition of MHC class II-restricted epitopes from NY-ESO-1 presented by a prevalent HLA DP4 allele: association with NY-ESO-1 antibody production. Proc Natl Acad Sci U S A 98:3964
Zeng G, Li Y, El Gamil M, Sidney J, Sette A, Wang RF, Rosenberg SA, Robbins PF (2002) Generation of NY-ESO-1-specific CD4+ and CD8+ T cells by a single peptide with dual MHC class I and class II specificities: a new strategy for vaccine design. Cancer Res 62:3630
Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213
Chen YT, Stockert E, Jungbluth A, Tsang S, Coplan KA, Scanlan MJ, Old LJ (1996) Serological analysis of Melan-A(MART-1), a melanocyte-specific protein homogeneously expressed in human melanomas. Proc Natl Acad Sci U S A 93:5915
Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, Snoke K, Serra HM, Kubo RT, Sette A (1994) Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1:751
Lang KS, Moris A, Gouttefangeas C, Walter S, Teichgraber V, Miller M, Wernet D, Hamprecht K, Rammensee HG, Stevanovic S (2002) High frequency of human cytomegalovirus (HCMV)-specific CD8+ T cells detected in a healthy CMV-seropositive donor. Cell Mol Life Sci 59:1076
Stockert E, Jager E, Chen YT, Scanlan MJ, Gout I, Karbach J, Arand M, Knuth A, Old LJ (1998) A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med 187:1349
Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A (1999) Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708
Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM (2000) NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res 60:4946
Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A (1998) Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 160:3363
Kwok WW, Gebe JA, Liu A, Agar S, Ptacek N, Hammer J, Koelle DM, Nepom GT (2001) Rapid epitope identification from complex class-II-restricted T-cell antigens. Trends Immunol 22:583
Knights AJ, Zaniou A, Rees RC, Pawelec G, Muller L (2002) Prediction of an HLA-DR-binding peptide derived from Wilms’ tumour 1 protein and demonstration of in vitro immunogenicity of WT1(124–138)-pulsed dendritic cells generated according to an optimised protocol. Cancer Immunol Immunother 51:271
Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, Trzeciak A, Gillessen D, Pink JR (1988) A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature 336:778
Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A (1989) Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol 19:2237
Kobayashi H, Wood M, Song Y, Appella E, Celis E (2000) Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res 60:5228
Romero P, Dutoit V, Rubio-Godoy V, Lienard D, Speiser D, Guillaume P, Servis K, Rimoldi D, Cerottini JC, Valmori D (2001) CD8+ T-cell response to NY-ESO-1: relative antigenicity and in vitro immunogenicity of natural and analogue sequences. Clin Cancer Res 7:766 s
Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM (2000) NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res 60:4946
Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM (2000) NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res 60:4946
Foukas PG, Tsilivakos V, Zacharatos P, Mariatos G, Moschos S, Syrianou A, Asimacopoulos PJ, Bramis J, Fotiadis C, Kittas C, Gorgoulis VG (2001) Expression of HLA-DR is reduced in tumor infiltrating immune cells (TIICs) and regional lymph nodes of non-small-cell lung carcinomas: a putative mechanism of tumor-induced immunosuppression? Anticancer Res 21:2609
Lazzaro B, Anderson AE, Kajdacsy-Balla A, Hessner MJ (2001) Antigenic characterization of medullary carcinoma of the breast: HLA-DR expression in lymph node positive cases. Appl Immunohistochem Mol Morphol 9:234
Ossendorp F, Mengede E, Camps M, Filius R, Melief CJ (1998) Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J Exp Med 187:693
Bennett SR, Carbone FR, Karamalis F, Miller JF, Heath WR (1997) Induction of a CD8+ cytotoxic T lymphocyte response by cross-priming requires cognate CD4+ T cell help. J Exp Med 186:65
Inaba K, Turley S, Yamaide F, Iyoda T, Mahnke K, Inaba M, Pack M, Subklewe M, Sauter B, Sheff D, Albert M, Bhardwaj N, Mellman I, Steinman RM (1998) Efficient presentation of phagocytosed cellular fragments on the major histocompatibility complex class II products of dendritic cells. J Exp Med 188:2163
Acknowledgements
We thank Claudia Schormann and Evi Regitz for excellent technical assistance, as well as Elisabeth Stockert (New York Branch of the LICR) and Elke Jäger (Krankenhaus Nordwest, Frankfurt/Main, Germany) for confirming the NY-ESO-1 serology data.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study was supported by BIOMED II (CT BMH4-C98-3589) of the European Commission, Pf-135/7-1, and by Kompetenznetz Maligne Lymphome (TP 11) of the BMBF.
Rights and permissions
About this article
Cite this article
Neumann, F., Wagner, C., Kubuschok, B. et al. Identification of an antigenic peptide derived from the cancer-testis antigen NY-ESO-1 binding to a broad range of HLA-DR subtypes. Cancer Immunol Immunother 53, 589–599 (2004). https://doi.org/10.1007/s00262-003-0492-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-003-0492-6