Abstract
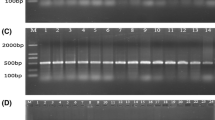
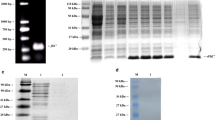
Salmonella spp. can cause animal and human salmonellosis. In this study, we established a simple method to detect all Salmonella species by amplifying a specific region within the flgE gene encoding the flagellar hook protein. Our preliminary sequence analysis among flagella-associated genes of Salmonella revealed that although Salmonella Gallinarum and Salmonella Pullorum are lacking flagella, they did have flagella-associated genes, including flgE. To investigate in detail, a comparative flgE sequence analysis was conducted using different bacterial strains including flagellated and non-flagellated Salmonella as well as non-Salmonella strains. Two unique regions (481–529 bp and 721–775 bp of the reference sequence) within the flgE open reading frame were found to be highly conserved and specific to all Salmonella species. Next, we designed a pair of PCR primers (flgE-UP and flgE-LO) targeting the above two regions, and performed a flgE-tailored PCR using as template DNA prepared from a total of 76 bacterial strains (31 flagellated Salmonella strains, 26 non-flagellated Salmonella strains, and 19 other non-Salmonella bacteria strains). Results showed that specific positive bands with expected size were obtained from all Salmonella (including flagellated and non-flagellated Salmonella) strains, while no specific product was generated from non-Salmonella bacterial strains. PCR products from the positive bands were confirmed by DNA sequencing. The minimum detection amount for genomic DNA and bacteria cells reached 18.3 pg/μL and 100 colony-forming unit (CFU) per PCR reaction, respectively. Using the flgE-PCR method to detect Salmonella in artificially contaminated milk samples, as low as 1 CFU/mL Salmonella was detectable after an 8-h pre-culture. Meanwhile, the flgE-tailored PCR method was applied to evaluate 247 clinical samples infected with Salmonella from different chicken breeding farms. The detection results indicated that flgE-PCR could be used to specifically detect Salmonella in concordance with the traditional bacterial culture-based detection method. It is worthwhile noticed that identification results using flgE-tailored PCR should be completed within less than 1 day, expanding the result of much faster than the standard method, which took more than 5 days. Overall, the flgE-tailored PCR method can specifically detect flagellated and non-flagellated Salmonella and can serve as a powerful tool for rapid, simple, and sensitive detection of Salmonella species.
Key points
• Targeting flgE gene for all Salmonella spp. found.
• The established PCR assay is used to specifically detect all Salmonella spp.
• The PCR method is applied to detect clinical Salmonella spp. samples within less than 1 day.







Similar content being viewed by others
References
Afroj S, Aldahami K, Reddy G, Guard J, Adesiyun A, Samuel T, Abdela W (2017) Simultaneous detection of multiple Salmonella serovars from milk and chicken meat by real-time PCR using unique genomic target regions. J Food Prot:1944–1957. https://doi.org/10.4315/0362-028X.JFP-17-133
Barrow PA, Freitas Neto OC (2011) Pullorum disease and fowl typhoid--new thoughts on old diseases: a review. Avian Pathol 40(1):1–13. https://doi.org/10.1080/03079457.2010.542575
Calvo L, Martinez-Planells A, Pardos-Bosch J, Garcia-Gil LJ (2008) A new real-time PCR assay for the specific detection of Salmonella spp. targeting the bipA gene. Food Anal Methods 1(4):236–242. https://doi.org/10.1007/s12161-007-9008-x
Chaban B, Hughes HV, Beeby M (2015) The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol 46:91–103. https://doi.org/10.1016/j.semcdb.2015.10.032
Chen S, Yee A, Griffiths M, Larkin C, Yamashiro CT, Behari R, Paszko-Kolva C, Rahn K, De Grandis SA (1997) The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol 35(3):239–250. https://doi.org/10.1016/s0168-1605(97)01241-5
Chen W, Martinez G, Mulchandani A (2000) Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem 280(1):166–172. https://doi.org/10.1006/abio.2000.4518
Chen J, Zhang L, Paoli GC, Shi C, Tu SI, Shi X (2010) A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int J Food Microbiol 137(2-3):168–174. https://doi.org/10.1016/j.ijfoodmicro.2009.12.004
Chiu TH, Pang JC, Hwang WZ, Tsen HY (2005) Development of PCR primers for the detection of Salmonella enterica serovar Choleraesuis based on the fliC gene. J Food Prot 68(8):1575–1580. https://doi.org/10.4315/0362-028x-68.8.1575
Cho IH, Ku S (2017) Current technical approaches for the early detection of foodborne pathogens: challenges and opportunities. Int J Mol Sci 18(10). https://doi.org/10.3390/ijms18102078
Cui BF, Liu XS, Fang YZ, Zhou P, Zhang YG, Wang YL (2018) Flagellin as a vaccine adjuvant. Expert Rev Vaccines 17(4):335–349. https://doi.org/10.1080/14760584.2018.1457443
de Almeida MV, Silva A Jr, Nero LA (2014) Evaluation of target sequences for the polymerase chain reaction-based detection of Salmonella in artificially contaminated beef. Foodborne Pathog Dis 11(2):111–118. https://doi.org/10.1089/fpd.2013.1623
Ding T, Suo Y, Zhang Z, Liu D, Ye X, Chen S, Zhao Y (2017) A multiplex RT-PCR assay for S. aureus, L. monocytogenes, and Salmonella spp. detection in raw milk with pre-enrichment. Front Microbiol 8:989. https://doi.org/10.3389/fmicb.2017.00989
Doran JL, Collinson SK, Burian J, Sarlos G, Todd EC, Munro CK, Kay CM, Banser PA, Peterkin PI, Kay WW (1993) DNA-based diagnostic tests for Salmonella species targeting agfA, the structural gene for thin, aggregative fimbriae. J Clin Microbiol 31(9):2263–2273
Duan Q, Zhou M, Zhu L, Zhu G (2013) Flagella and bacterial pathogenicity. J Basic Microbiol 53(1):1–8. https://doi.org/10.1002/jobm.201100335
Foley SL, Lynne AM (2008) Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci 86(14 Suppl):E173–E187. https://doi.org/10.2527/jas.2007-0447
Gupta SK, Bajwa P, Deb R, Chellappa MM, Dey S (2014) Flagellin a Toll-like receptor 5 agonist as an adjuvant in chicken vaccines. Clin Vaccine Immunol 21(3):261–270. https://doi.org/10.1128/Cvi.00669-13
Hadjinicolaou AV, Demetriou VL, Emmanuel MA, Kakoyiannis CK, Kostrikis LG (2009) Molecular beacon-based real-time PCR detection of primary isolates of Salmonella Typhimurium and Salmonella Enteritidis in environmental and clinical samples. BMC Microbiol 9:97. https://doi.org/10.1186/1471-2180-9-97
Hajam IA, Dar PA, Shahnawaz I, Jaume JC, Lee JH (2017) Bacterial flagellin-a potent immunomodulatory agent. Exp Mol Med 49(9):e373. https://doi.org/10.1038/emm.2017.172
Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410(6832):1099–1103. https://doi.org/10.1038/35074106
Hirose K, Itoh K, Nakajima H, Kurazono T, Yamaguchi M, Moriya K, Ezaki T, Kawamura Y, Tamura K, Watanabe H (2002) Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. J Clin Microbiol 40(2):633–636. https://doi.org/10.1128/jcm.40.02.633-636.2002
Honko AN, Mizel SB (2005) Effects of flagellin on innate and adaptive immunity. Immunol Res 33(1):83–101. https://doi.org/10.1385/IR:33:1:083
Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, Nair S, Fields PI, Weill FX (2014) Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res Microbiol 165(7):526–530. https://doi.org/10.1016/j.resmic.2014.07.004
Khan S, Harish BN, Menezes GA, Acharya NS, Parija SC (2012) Early diagnosis of typhoid fever by nested PCR for flagellin gene of Salmonella enterica serotype Typhi. Indian J Med Res 136(5):850–854
Kim MI, Lee C, Park J, Jeon BY, Hong M (2018) Structure-guided fusion-protein designs using Bacillus flagellin as a vaccine adjuvant. Acta Crystallogr A 74:A134–A134. https://doi.org/10.1107/S0108767318098653
Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ (2015) World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12(12):e1001921. https://doi.org/10.1371/journal.pmed.1001921
Kwang J, Littledike ET, Keen JE (1996) Use of the polymerase chain reaction for Salmonella detection. Lett Appl Microbiol 22(1):46–51. https://doi.org/10.1111/j.1472-765x.1996.tb01106.x
Li Y, Shen Y, Lin D, Zhang H, Wang T, Liu H, Wang Y (2019) Neutrophils and IL17A mediate flagellar hook protein FlgE-induced mouse acute lung inflammation. Cell Microbiol 21(3):e12975. https://doi.org/10.1111/cmi.12975
Lin CK, Hung CL, Hsu SC, Tsai CC, Tsen HY (2004) An improved PCR primer pair based on 16S rDNA for the specific detection of Salmonella serovars in food samples. J Food Prot 67(7):1335–1343. https://doi.org/10.4315/0362-028x-67.7.1335
Makino S, Kurazono H, Chongsanguam M, Hayashi H, Cheun H, Suzuki S, Shirahata T (1999) Establishment of the PCR system specific to Salmonella spp. and its application for the inspection of food and fecal samples. J Vet Med Sci 61(11):1245–1247. https://doi.org/10.1292/jvms.61.1245
Malorny B, Paccassoni E, Fach P, Bunge C, Martin A, Helmuth R (2004) Diagnostic real-time PCR for detection of Salmonella in food. Appl Environ Microbiol 70(12):7046–7052. https://doi.org/10.1128/AEM.70.12.7046-7052.2004
Malorny B, Huehn S, Dieckmann R, Krmer N, Helmuth R (2009) Polymerase chain reaction for the rapid detection and serovar identification of Salmonella in food and feeding stuff. Food Anal Methods 2(2):81–95. https://doi.org/10.1007/s12161-008-9057-9
Matsunami H, Barker CS, Yoon YH, Wolf M, Samatey FA (2016) Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat Commun 7:13425. https://doi.org/10.1038/ncomms13425
Moriya N, Minamino T, Imada K, Namba K (2011) Genetic analysis of the bacterial hook-capping protein FlgD responsible for hook assembly. Microbiol-SGM 157:1354–1362. https://doi.org/10.1099/mic.0.047100-0
Munir T, Lodhi M, Ali S, Hussain Zaidi SB, Razak S (2015) Early diagnosis of typhoid by PCR for fliC-d gene of Salmonella Typhi in patients taking antibiotics. J Coll Physicians Surg Pak 25(9):662–666
Nam HM, Srinivasan V, Gillespie BE, Murinda SE, Oliver SP (2005) Application of SYBR green real-time PCR assay for specific detection of Salmonella spp. in dairy farm environmental samples. Int J Food Microbiol 102(2):161–171. https://doi.org/10.1016/j.ijfoodmicro.2004.12.020
Pathmanathan SG, Cardona-Castro N, Sanchez-Jimenez MM, Correa-Ochoa MM, Puthucheary SD, Thong KL (2003) Simple and rapid detection of Salmonella strains by direct PCR amplification of the hilA gene. J Med Microbiol 52(Pt 9):773–776. https://doi.org/10.1099/jmm.0.05188-0
Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, Curtiss R 3rd, Gyles CL (1992) Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 6(4):271–279. https://doi.org/10.1016/0890-8508(92)90002-f
Ricke SC, Kim SA, Shi Z, Park SH (2018) Molecular-based identification and detection of Salmonella in food production systems: current perspectives. J Appl Microbiol 125(2):313–327. https://doi.org/10.1111/jam.13888
Rukambile E, Sintchenko V, Muscatello G, Kock R, Alders R (2019) Infection, colonization and shedding of Campylobacter and Salmonella in animals and their contribution to human disease: a review. Zoonoses Public Health 66(6):562–578. https://doi.org/10.1111/zph.12611
Saijo-Hamano Y, Matsunami H, Namba K, Imada K (2019) Architecture of the bacterial flagellar distal rod and hook of Salmonella. Biomole 9(7). https://doi.org/10.3390/biom9070260
Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, Derosier DJ, Kitao A, Namba K (2004) Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431(7012):1062–1068. https://doi.org/10.1038/nature02997
Shen Y, Chen L, Wang M, Lin D, Liang Z, Song P, Yuan Q, Tang H, Li W, Duan K, Liu B, Zhao G, Wang Y (2017) Flagellar hooks and hook protein FlgE participate in host microbe interactions at immunological level. Sci Rep 7(1):1433. https://doi.org/10.1038/s41598-017-01619-1
Threlfall EJ (2002) Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol Rev 26(2):141–148. https://doi.org/10.1111/j.1574-6976.2002.tb00606.x
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Ye XH, Wang YM, Lin XG (2011) A gyrB-targeted PCR for rapid identification of Salmonella. Curr Microbiol 63(5):477–483. https://doi.org/10.1007/s00284-011-0007-1
Zhang JY, Dong LW, Ren Q, Wang XZ, Yang Y, Zhou W, Zhu CH, Meng X, Zhu GQ (2014) Simple and rapid detection of Salmonella by direct PCR amplification of gene fimW. Curr Microbiol 69(4):429–435. https://doi.org/10.1007/s00284-014-0602-z
Zhou MX, Yang Y, Chen PL, Hu HJ, Hardwidge PR, Zhu GQ (2015) More than a locomotive organelle: flagella in Escherichia coli. Appl Microbiol Biotechnol 99(21):8883–8890. https://doi.org/10.1007/s00253-015-6946-x
Zhou B, Liang T, Zhan Z, Liu R, Li F, Xu H (2017) Rapid and simultaneous quantification of viable Escherichia coli O157:H7 and Salmonella spp. in milk through multiplex real-time PCR. J Dairy Sci 100(11):8804–8813. https://doi.org/10.3168/jds.2017-13362
Acknowledgments
The authors gratefully acknowledge Professor Xinan Jiao in Yangzhou University, Professor Chengping Lu in Nanjing Agricultural University, Professor Shulin Liu from Harbin Medical University and the Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences for providing the bacteria strains for this study.
Funding
This work was supported by the grants from the no. 2016YFD0500905 and 2017YFD0500105 from the National Key Research and Development Program of China, and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
YY and PW performed the experiments, analyzed the data, and wrote the manuscript. PX, BY, PD, TH, and JL participated in the data analysis and wrote the paper. QS contributed to the experiments designing, manuscript writing, and language polishing. GZ and XM conceived and designed the study, participated in experimental work, and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This article does not contain any studies with human participants performed by any of the authors. The current study was approved by the by the Institutional Animal Care and Use Committee of the Yangzhou University College of Veterinary Medicine of China.
Consent for publication
All authors listed on this manuscript have read and agreed to the publication of this research.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 3078 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Wang, P., Xia, P. et al. Rapid detection of flagellated and non-flagellated Salmonella by targeting the common flagellar hook gene flgE. Appl Microbiol Biotechnol 104, 9719–9732 (2020). https://doi.org/10.1007/s00253-020-10925-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10925-0