Abstract
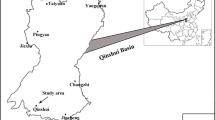
To identify the methanogenic pathways present in a deep coal bed methane (CBM) reservoir associated with Eastern Ordos Basin in China, a series of geochemical and microbiological studies was performed using gas and water samples produced from the Liulin CBM reservoir. The composition and stable isotopic ratios of CBM implied a mixed biogenic and thermogenic origin of the methane. Archaeal 16S rRNA gene analysis revealed the dominance of the methylotrophic methanogen Methanolobus in the water produced. The high potential of methane production by methylotrophic methanogens was found in the enrichments using the water samples amended with methanol and incubated at 25 and 35 °C. Methylotrophic methanogens were the dominant archaea in both enrichments as shown by polymerase chain reaction (PCR)–denaturing gradient gel electrophoresis (DGGE). Bacterial 16S rRNA gene analysis revealed that fermentative, sulfate-reducing, and nitrate-reducing bacteria inhabiting the water produced were a factor in coal biodegradation to fuel methanogens. These results suggested that past and ongoing biodegradation of coal by methylotrophic methanogens and syntrophic bacteria, as well as thermogenic CBM production, contributed to the Liulin CBM reserves associated with the Eastern Ordos Basin.






Similar content being viewed by others
References
Allen TD, Caldwell ME, Lawson PA, Huhnke RL, Tanner RS (2010) Alkalibaculum bacchi gen. nov., sp. nov., a CO-oxidizing, ethanol-producing acetogen isolated from livestock-impacted soil. Int J Syst Evol Microbiol 60:2483–2489
Bates BL, McIntosh JC, Lohse KA, Brooks PD (2011) Influence of groundwater flowpaths, residence times and nutrients on the extent of microbial methanogenesis in coal beds: Powder River Basin, USA. Chem Geol 284:45–61
Bernard BB, Brooks JM, Sackett WM (1978) Light hydrocarbons in recent Texas continental shelf and slope sediments. J Geophys Res 83:4053–4061
Chen S, Niu L, Zhang Y (2010) Saccharofermentans acetigenes gen nov, sp nov, an anaerobic bacterium isolated from sludge treating brewery wastewater. Int J Syst Evol Microbiol 60:2735–2738
Chou JH, Jiang SR, Cho JC, Song J, Lin MC, Chen WM (2008) Azonexus hydrophilus sp nov, a nifH gene-harbouring bacterium isolated from freshwater. Int J Syst Evol Microbiol 58:946–951
Collado L, Levican A, Perez J, Figueras MJ (2011) Arcobacter defluvii sp nov, isolated from sewage samples. Int J Syst Evol Microbiol 61:2155–2161
Cuzin N, Ouattara AS, Labat M, Garcia JL (2001) Methanobacterium congolense sp nov, from a methanogenic fermentation of cassava peel. Int J Syst Evol Microbiol 51:489–493
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89:5685–5689
Ding S, Zhu J, Zhen B, Liu Z, Liu Q (2011) Characteristics of high rank coal bed methane reservoir from the Xiangning Mining Area, Eastern Ordos Basin, China. Energ Explor Exploit 29:33–48
Donnelly M, Dagley S (1980) Production of methanol from aromatic acids by Pseudomonas putida. J Bacteriol 142:916–924
Flores RM, Rice CA, Stricker GD, Warden A, Ellis MS (2008) Methanogenic pathways of coal-bed gas in the Powder River Basin, United States: the geologic factor. Int J Coal Geol 76:52–75
Friedrich M, Springer N, Ludwig W, Schink B (1996) Phylogenetic positions of Desulfofustis glycolicus gen. nov., sp. nov. and Syntrophobotulus glycolicus gen. nov., sp. nov., two new strict anaerobes growing with glycolic acid. Int J Syst Evol Microbiol 46:1065–1069
Garnova ES, Zhilina TN, Tourova TP, Kostrikina NA, Zavarzin GA (2004) Anaerobic, alkaliphilic, saccharolytic bacterium Alkalibacter saccharofermentans gen. nov., sp. nov. from a soda lake in the Transbaikal region of Russia. Extremophiles 8:309–316
Green MS, Flanegan KC, Gilcrease PC (2008) Characterization of a methanogenic consortium enriched from a coal bed methane well in the Powder River Basin, USA. Int J Coal Geol 76:34–45
Gunsalus RP, Romesser JA, Wolfe RS (1978) Preparation of coenzyme M analogs and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry 17:2374–2377
Harris SH, Smith RL, Barker CE (2008) Microbial and chemical factors influencing methane production in laboratory incubations of low-rank subsurface coals. Int J Coal Geol 76:46–51
Hippe H, Vainshtein M, Gogotova G, Stackebrandt E (2003) Reclassification of Desulfobacterium macestii as Desulfomicrobium macestii comb. nov. Int J Syst Evol Microbiol 53:1127–1130
Kato S, Haruta S, Cui ZJ, Ishii M, Yokota A, Igarashi Y (2004) Clostridium straminisolvens sp. nov., a moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int J Syst Evol Microbiol 54:2043–2047
Krumholz LR, Harris SH, Tay ST, Suflita JM (1999) Characterization of two subsurface H-2-utilizing bacteria, Desulfomicrobium hypogeium sp nov and Acetobacterium psammolithicum sp nov., and their ecological roles. Appl Environ Microbiol 65:2300–2306
Lane D (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Liu A, Garcia-Dominguez E, Rhine E, Young L (2004) A novel arsenate respiring isolate that can utilize aromatic substrates. FEMS Microbiol Ecol 48:323–332
Matsutani N, Nakagawa T, Nakamura K, Takahashi R, Yoshihara K, Tokuyama T (2009) Enrichment of a novel marine ammonia-oxidizing archaeon obtained from sand of an eelgrass zone. Microb Environ 26:23–29
Mechichi T, Stackebrandt E, Gad'on N, Fuchs G (2002) Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch Microbiol 178:26–35
Midgley DJ, Hendry P, Pinetown KL, Fuentes D, Gong S, Mitchell DL, Faiz M (2010) Characterisation of a microbial community associated with a deep, coal seam methane reservoir in the Gippsland Basin, Australia. Int J Coal Geol 82:232–239
Mochimaru H, Tamaki H, Hanada S, Imachi H, Nakamura K, Sakata S, Kamagata Y (2009) Methanolobus profundi sp. nov., a methylotrophic methanogen isolated from deep subsurface sediments in a natural gas field. Int J Syst Evol Microbiol 59:714–718
Morasch B, Schink B, Tebbe CC, Meckenstock RU (2004) Degradation of o-xylene and m-xylene by a novel sulfate-reducer belonging to the genus Desulfotomaculum. Arch Microbiol 181:407–417
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Oremland RS, Polcin S (1982) Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl Environ Microbiol 44:1270–1276
Paarup M, Friedrich MW, Tindall BJ, Finster K (2006) Characterization of the psychrotolerant acetogen strain SyrA5 and the emended description of the species Acetobacterium carbinolicum. Anton Leeuw 89:55–69
Penner T, Foght J, Budwill K (2010) Microbial diversity of western Canadian subsurface coal beds and methanogenic coal enrichment cultures. Int J Coal Geol 82:81–93
Pfeiffer RS, Ulrich GA, Finkelstein M (2010) Chemical amendments for the stimulation of biogenic gas generation in deposits of carbonaceous material. US Patent No. 7696132
Qiu YL, Sekiguchi Y, Imachi H, Kamagata Y, Tseng IC, Cheng SS, Ohashi A, Harada H (2004) Identification and isolation of anaerobic, syntrophic phthalate isomer-degrading microbes from methanogenic sludges treating wastewater from terephthalate manufacturing. Appl Environ Microbiol 70:1617–1626
Ramamoorthy S, Sass H, Langner H, Schumann P, Kroppenstedt R, Spring S, Overmann J, Rosenzweig R (2006) Desulfosporosinus lacus sp. nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. Int J Syst Evol Microbiol 56:2729–2736
Raskin L, Stromley J, Rittmann B, Stahl D (1994) Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60:1232–1240
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Scott AR, Kaiser W, Ayers WB (1994) Thermogenic and secondary biogenic gases, San Juan Basin, Colorado and New Mexico—implications for coal bed gas producibility. Bull Am Assoc Petrol Geol 78:1186–1209
Shimizu S, Akiyama M, Naganuma T, Fujioka M, Nako M, Ishijima Y (2007) Molecular characterization of microbial communities in deep coal seam groundwater of northern Japan. Geobiology 5:423–433
Shimizu S, Upadhye R, Ishijima Y, Naganuma T (2011) Methanosarcina horonobensis sp. nov., a methanogenic archaeon isolated from a deep subsurface Miocene formation. Int J Syst Evol Microbiol 61:2503–2507
Shinoda Y, Sakai Y, Uenishi H, Uchihashi Y, Hiraishi A, Yukawa H, Yurimoto H, Kato N (2004) Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl Environ Microbiol 70:1385–1392
Simankova MV, Kotsyurbenko OR, Lueders T, Nozhevnikova AN, Wagner B, Conrad R, Friedrich MW (2003) Isolation and characterization of new strains of methanogens from cold terrestrial habitats. Syst Appl Microbiol 26:312–318
Smith CJ, Nedwell DB, Dong LF, Osborn AM (2006) Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ Microbiol 8:804–815
Steven B, Briggs G, McKay CP, Pollard WH, Greer CW, Whyte LG (2007) Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol Ecol 59:513–523
Strapoć D, Mastalerz M, Eble C, Schimmelmann A (2007) Characterization of the origin of coal bed gases in southeastern Illinois Basin by compound-specific carbon and hydrogen stable isotope ratios. Org Geochem 38:267–287
Strapoć D, Picardal FW, Turich C, Schaperdoth I, Macalady JL, Lipp JS, Lin YS, Ertefai TF, Schubotz F, Hinrichs KU, Mastalerz M, Schimmelmann A (2008) Methane-producing microbial community in a coal bed of the Illinois Basin. Appl Environ Microbiol 74:2424–2432
Strapoć D, Ashby M, Wood L, Levinson R, Huizinga B (2010) Significant contribution of methyl/methanol-utilising methanogenic pathway in a subsurface biogas environment. In: Skovhus T, Whitby C (eds) Applied microbiology and molecular biology in oilfield systems. Springer, pp 211–216
Strapoć D, Mastalerz M, Dawson K, Macalady J, Callaghan AV, Wawrik B, Turich C, Ashby M (2011) Biogeochemistry of microbial coal-bed methane. Annu Rev Earth Pl Sc 39:617–656
Su X, Zhang L, Zhang R (2003) The abnormal pressure regime of the Pennsylvanian No. 8 coal bed methane reservoir in Liulin-Wupu District, Eastern Ordos Basin, China. Int J Coal Geol 53:227–239
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wu SY, Lai MC (2011) Methanogenic archaea isolated from Taiwan’s Chelungpu Fault. Appl Environ Microbiol 77:830–838
Zhang G, Jiang N, Liu X, Dong X (2008) Methanogenesis from methanol at low temperatures by a novel psychrophilic methanogen, “Methanolobus psychrophilus” sp. nov., prevalent in Zoige Wetland of the Tibetan Plateau. Appl Environ Microbiol 74:6114–6120
Zhilina T, Kevbrin V, Tourova T, Lysenko A, Kostrikina N, Zavarzin G (2005) Clostridium alkalicellum sp. nov., an obligately alkaliphilic cellulolytic bacterium from a soda lake in the Baikal region. Microbiology 74:557–566
Acknowledgements
We are grateful to China United Coalbed Methane Co., Ltd., for helping with sampling opportunities on the water produced. This work was supported by National Science and Technology Major Project, China (2011ZX05060-005 and 2009ZX05039-003).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Guo, H., Yu, Z., Liu, R. et al. Methylotrophic methanogenesis governs the biogenic coal bed methane formation in Eastern Ordos Basin, China. Appl Microbiol Biotechnol 96, 1587–1597 (2012). https://doi.org/10.1007/s00253-012-3889-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-3889-3