Abstract
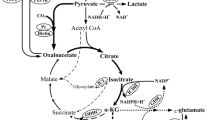
We recently showed that the activity of the 2-oxoglutarate dehydrogenase complex (ODHC) in Corynebacterium glutamicum is controlled by a novel regulatory mechanism that involves a 15-kDa protein called OdhI and serine/threonine protein kinase G (PknG). In its unphosphorylated state, OdhI binds to the E1 subunit (OdhA) of ODHC and, thereby, inhibits its activity. Inhibition is relieved by phosphorylation of OdhI at threonine-14 by PknG under conditions requiring high ODHC activity. In this work, evidence is provided that the dephosphorylation of phosphorylated OdhI is catalyzed by a phospho-Ser/Thr protein phosphatase encoded by the gene cg0062, designated ppp. As a decreased ODHC activity is important for glutamate synthesis, we investigated the role of OdhI and PknG for glutamate production under biotin limitation and after addition of Tween-40, penicillin, or ethambutol. A ΔodhI mutant formed only 1–13% of the glutamate synthesized by the wild type. Thus, OdhI is essential for efficient glutamate production. The effect of a pknG deletion on glutamate synthesis was dependent on the induction conditions. Under strong biotin limitation and in the presence of ethambutol, the ΔpknG mutant showed significantly increased glutamate production, offering a new way to improve production strains.



Similar content being viewed by others
Abbreviations
- cdw:
-
cell dry weight
- PknG:
-
serine/threonine protein kinase G
- ODHC:
-
2-oxoglutarate dehydrogenase complex
- OdhI:
-
oxoglutarate dehydrogenase inhibitor protein
- TCA cycle:
-
tricarboxylic acid cycle
References
Asakura Y, Kimura E, Usuda Y, Kawahara Y, Matsui K, Osumi T, Nakamatsu T (2007) Altered metabolic flux due to deletion of odhA causes l-glutamate overproduction in Corynebacterium glutamicum. Appl Environ Microbiol 73:1308–1319
Delaunay S, Gourdon P, Lapujade P, Mailly E, Oriol E, Engasser JM, Lindley ND, Goergen JL (1999) An improved temperature triggered process for glutamate production with Corynebacterium glutamicum. Enzyme Microb Technol 25:762–768
Duperray F, Jezequel D, Ghazi A, Letellier L, Shechter, E. (1992) Excretion of glutamate from Corynebacterium glutamicum triggered by amine surfactants. Biochim Biophys Acta 1103:250–258
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL, USA
Eggeling L, Krumbach K, Sahm H (2001) l-glutamate efflux with Corynebacterium glutamicum: why is penicillin treatment or Tween addition doing the same? J Mol Microbiol Biotechnol 3:67–68
Gutmann M, Hoischen C, Krämer R (1992) Carrier-mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim Biophys Acta 1112:115–123
Hanahan D (1985) Techniques for transformation of E. coli. In: Glover DM (eds) DNA-Cloning: a practical approach. IRL-Press, Oxford, pp 109–135
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Hoischen C, Krämer R (1989) Evidence for an efflux carrier system involved in the secretion of glutamate by Corynebacterium glutamicum. Arch Microbiol 151:342–347
Hoischen C, Krämer R (1990) Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J Bacteriol 172:3409–3416
Kabus A, Niebisch A, Bott M (2007) Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl Environ Microbiol 73:861–868
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Kanzaki T, Isobe K, Okazaki H, Motizuki K, Fukuda H (1967) l-Glutamic acid production. Part I. Selection of an oleic acid-requiring mutant and its properties. Agric Biol Chem 31:1307–1313
Kataoka M, Hashimoto KI, Yoshida M, Nakamatsu T, Horinouchi S, Kawasaki H (2006) Gene expression of Corynebacterium glutamicum in response to the conditions inducing glutamate overproduction. Lett Appl Microbiol 42:471–476
Kawahara Y, Takahashi-Fuke K, Shimizu E, Nakamatsu T, Nakamori S (1997) Relationship between the glutamate production and the activity of 2-oxoglutarate dehydrogenase in Brevibacterium lactofermentum. Biosci Biotechnol Biochem 61:1109–1112
Keilhauer C, Eggeling L, Sahm H (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB–ilvN–ilvC operon. J Bacteriol 175:5595–5603
Kimura E (2002) Triggering mechanism of l-glutamate overproduction by DtsR1 in coryneform bacteria. J Biosci Bioeng 94:545–551
Kimura E, Abe C, Kawahara Y, Nakamatsu T, Tokuda H (1997) A dtsR gene-disrupted mutant of Brevibacterium lactofermentum requires fatty acids for growth and efficiently produces l-glutamate in the presence of an excess of biotin. Biochem Biophys Res Commun 234:157–161
Kinoshita S, Udaka S, Shimono M (1957) Studies on the amino acid fermentation. I. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol 3:193–205
Krug A, Wendisch VF, Bott M (2005) Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J Biol Chem 280:585–595
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Liebl W (2005) Corynebacterium taxonomy. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, pp 9–34
Momose H, Takagi T (1978) Glutamic acid production in biotin-rich media by temperature sensitive mutants of Brevibacterium lactofermentum, a novel fermentation process. Agric Biol Chem 42:1911–1917
Nakao Y, Kikuchi M, Suzuki M, Doi M (1972) Microbial production of l-glutamic acid by glycerol auxotrophs. I. Induction of glycerol auxotrophs and production of l-glutamic acid from n-paraffins. Agric Biol Chem 36:490–496
Nampoothiri KM, Hoischen C, Bathe B, Möckel B, Pfefferle W, Krumbach K, Sahm H, Eggeling L (2002) Expression of genes of lipid synthesis and altered lipid composition modulates l-glutamate efflux of Corynebacterium glutamicum. Appl Microbiol Biotechnol 58:89–96
Niebisch A, Bott M (2001) Molecular analysis of the cytochrome bc 1–aa 3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c 1. Arch Microbiol 175:282–294
Niebisch A, Kabus A, Schultz C, Weil B, Bott M (2006) Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J Biol Chem 281:12300–12307
Nunheimer TD, Birnbaum J, Ihnen ED, Demain AL (1970) Product inhibition of the fermentative formation of glutamic acid. Appl Microbiol 20:215–217
Okazaki H, Kanzaki T, Doi M, Sumino Y, Fukuda H (1967) l-Glutamic acid fermentation. Part II. The production of l-glutamic acid by an oleic acid-requiring mutant. Agric Biol Chem 31:1314–1317
Radmacher E, Stansen KC, Besra GS, Alderwick LJ, Maughan WN, Hollweg G, Sahm H, Wendisch VF, Eggeling L (2005) Ethambutol, a cell wall inhibitor of Mycobacterium tuberculosis, elicits l-glutamate efflux of Corynebacterium glutamicum. Microbiology 151:1359–1368
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Shiio I, Ujigawa-Takeda K (1980) Presence and regulation of α-ketoglutarate dehydrogenase complex in a glutamate producing bacterium, Brevibacterium flavum. Agric Biol Chem 44:1897–1904
Shiio I, Otsuka SI, Takahashi M (1962a) Effect of biotin on the bacterial formation of glutamic acid. I. Glutamate formation and cellular permeability of amino acids. J Biochem 51:56–62
Shiio I, Otsuka SI, Katsuya N (1962b) Effect of biotin on the bacterial formation of glutamic acid. II. Metabolism of glucose. J Biochem 52:108–116
Shimizu H, Tanaka H, Nakato A, Nagahisa K, Kimura E, Shioya S (2003) Effects of the changes in enzyme activities on metabolic flux redistribution around the 2-oxoglutarate branch in glutamate production by Corynebacterium glutamicum. Bioprocess Biosyst Eng 25:291–298
Shingu H, Terui G (1971) Studies on the process of glutamic acid fermentation at the enzyme level: I. On the changes of α-ketoglutaric acid dehydrogenase in the course of culture. J Ferment Technol 49:400–405
Shirai T, Nakato A, Izutani N, Nagahisa K, Shioya S, Kimura E, Kawarabayasi Y, Yamagishi A, Gojobori T, Shimizu H (2005) Comparative study of flux redistribution of metabolic pathway in glutamate production by two coryneform bacteria. Metab Eng 7:59–69
Takinami K, Yoshii H, Tsuri H, Okada H (1965) Biochemical effects of fatty acid and its derivatives on l-glutamic acid fermentation: III. Biotin–Tween 60 relationship in the accumulation of l-glutamic acid and the growth of Brevibacterium lactofermentum. Agric Biol Chem 29:351–359
Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Usuda Y, Tujimoto N, Abe C, Asakura Y, Kimura E, Kawahara Y, Kurahashi O, Matsui H (1996) Molecular cloning of the Corynebacterium glutamicum (‘Brevibacterium lactofermentum’ AJ12036) odhA gene encoding a novel type of 2-oxoglutarate dehydrogenase. Microbiology 142:3347–3354
Uy D, Delaunay S, Goergen JL, Engasser JM (2005) Dynamics of glutamate synthesis and excretion fluxes in batch and continuous cultures of temperature-triggered Corynebacterium glutamicum. Bioprocess Biosyst Eng 27:153–162
van der Rest ME, Lange C, Molenaar D (1999) A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52:541–545
Villarino A, Duran R, Wehenkel A, Fernandez P, England P, Brodin P, Cole ST, Zimny-Arndt U, Jungblut PR, Cervenansky C, Alzari PM (2005) Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J Mol Biol 350:953–963
Wennerhold J, Bott M (2006) The DtxR regulon of Corynebacterium glutamicum. J Bacteriol 188:2907–2918
Wennerhold J, Krug A, Bott M (2005) The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem 280:40500–40508
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to Prof. Dr. Hermann Sahm on the occasion of his 65th birthday
Rights and permissions
About this article
Cite this article
Schultz, C., Niebisch, A., Gebel, L. et al. Glutamate production by Corynebacterium glutamicum: dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl Microbiol Biotechnol 76, 691–700 (2007). https://doi.org/10.1007/s00253-007-0933-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-0933-9