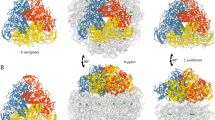
Abstract
The protein 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN) is involved in the quorum sensing of several bacterial species, including Helicobacter pylori. In particular, these bacteria depend on MTAN for synthesis of vitamin K2 homologs. The residue D198 in the active site of MTAN seems to be of crucial importance, by acting as a hydrogen-bond acceptor for the ligand. In this study, we investigated the conformation and dynamics of apo and holo H. pylori MTAN (HpMTAN), and assessed the effect of protonation of D198 by use of molecular dynamics simulations. Our results show that protonation of the active site of HpMTAN can cause a conformational transition from a closed state to an open state even in the absence of substrate, via inter-chain mechanical coupling.









Similar content being viewed by others
References
Appleby TC, Erion MD, Ealick SE (1999) The structure of human 5′-deoxy-5′-methylthioadenosine phosphorylase at 1.7 A resolution provides insights into substrate binding and catalysis. Structure 7:629–641
Bao Y, Li Y, Jiang Q, Zhao L, Xue T, Hu B, Sun B (2013) Methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs) of Staphylococcus aureus is essential for the virulence independent of LuxS/AI-2 system. Int J Med Microbiol 303:190–200
Bardhan PK (1997) Epidemiological features of Helicobacter pylori infection in developing countries. Clin Infect Dis 25:973–978
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:14101
da Silva AS, Vranken W (2012) ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res Notes 5:1–8
Darden T, York D, Pedersen L (1993) Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Dooley CP, Cohen H, Fitzgibbons PL, Bauer M, Appleman MD, Perezperez GI, Blaser MJ (1989) Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med 321:1562–1566
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. Journal of Chemical Physics 103:8577–8593
Evans GB, Fumeaux RH, Greatrex B, Murkin AS, Schramm VL, Tyler PC (2008) Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem 51:948–956
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Laham A, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, Revision C.02
Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275
Gutierrez JA, Luo M, Singh V, Li L, Brown RL, Norris GE, Evans GB, Furneaux RH, Tyler PC, Painter GF, Lenz DH, Schramm VL (2007) Picomolar inhibitors as transition-state probes of 5′-methylthioadenosine nucleosidases. ACS Chem Biol 2:725–734
Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL (2009) Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat Chem Biol 5:251–257
Haapalainen AM, Thomas K, Tyler PC, Evans GB, Almo SC, Schramm VL (2013) Salmonella enterica MTAN at 1.36 A resolution: a structure-based design of tailored transition state analogs. Structure 21:963–974
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T (2008) An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321:1670–1673
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(33–38):27–38
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23:1623–1641
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Kashiwagi H (2003) Ulcers and gastritis. Endoscopy 35:9–14
Lee JE, Cornell KA, Riscoe MK, Howell PL (2001) Structure of E. coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases. Structure 9:941–953
Lee JE, Cornell KA, Riscoe MK, Howell PL (2003) Structure of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. J Biol Chem 278:8761–8770
Lee JE, Singh V, Evans GB, Tyler PC, Furneaux RH, Cornell KA, Riscoe MK, Schramm VL, Howell PL (2005) Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. J Biol Chem 280:18274–18282
Li X, Apel D, Gaynor EC, Tanner ME (2011) 5′-Methylthioadenosine nucleosidase is implicated in playing a key role in a modified futalosine pathway for menaquinone biosynthesis in Campylobacter jejuni. J Biol Chem 286:19392–19398
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958
Longshaw AI, Adanitsch F, Gutierrez JA, Evans GB, Tyler PC, Schramm VL (2010) Design and synthesis of potent “sulfur-free” transition state analogue inhibitors of 5′-methylthioadenosine nucleosidase and 5′-methylthioadenosine phosphorylase. J Med Chem 53:6730–6746
Marshall BJ, Warren JR (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic-ulceration. Lancet 1:1311–1315
Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199
Mishra V, Ronning DR (2012) Crystal structures of the Helicobacter pylori MTAN enzyme reveal specific interactions between S-adenosylhomocysteine and the 5′-alkylthio binding subsite. Biochemistry 51:9763–9772
Mishra V, Ronning DR (2013) Enzyme-substrate interactions in the Helicobacter pylori 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. FASEB J 27:560–572
Parrinello M, Rahman A (1981) Polymorphic transitions in single-crystals—a new molecular-dynamics method. J Appl Phys 52:7182–7190
Parveen N, Cornell KA (2011) Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol 79:7–20
Plummer M, Franceschi S, Munoz N (2004) Epidemiology of gastric cancer. IARC Sci Publ 157:311–326
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Ronning DR, Iacopelli NM, Mishra V (2010) Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Protein Sci 19:2498–2510
Salomon-Ferrer R, Case DA, Walker RC (2013) An overview of the Amber biomolecular simulation package. Wiley Interdiscip Rev: Comput Mol Sci 3:198–210
Schrodinger, LLC (2010) The PyMOL Molecular Graphics System, Version 1.3r1
Seto H, Jinnai Y, Hiratsuka T, Fukawa M, Furihata K, Itoh N, Dairi T (2008) Studies on a new biosynthetic pathway for menaquinone. J Am Chem Soc 130:5614–5615
Singh V, Schramm VL (2007) Transition-state analysis of S-pneumoniae 5′-methylthioadenosine nucleosidase. J Am Chem Soc 129:2783–2795
Sipponen P (1997) Helicobacter pylori gastritiS-epidemiology. J Gastroenterol 32:273–277
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Wang SZ, Haapalainen AM, Yan FN, Du Q, Tyler PC, Evans GB, Rinaldo-Matthis A, Brown RL, Norris GE, Almo SC, Schramm VL (2012) A picomolar transition state analogue inhibitor of MTAN as a Specific antibiotic for Helicobacter pylori. Biochemistry 51:6892–6894
Wang S, Thomas K, Schramm VL (2014) Catalytic site cooperativity in dimeric methylthioadenosine nucleosidase. Biochemistry 53:1527–1535
Wassenaar TA, Quax WJ, Mark AE (2008) The conformation of the extracellular binding domain of death receptor 5 in the presence and absence of the activating ligand TRAIL: a molecular dynamics study. Proteins 70:333–343
Wold H (1966) Estimation of principal component and related models by iterative least squares. Multivariate Analysis. Academic Press, New York, pp 391–420
Yildirim A, Tekpinar M, Wassenaar TA (2014) Molecular dynamics investigation of Helicobacter pylori chemotactic protein CheY1 and two mutants. J Mol Model 20:2212
Acknowledgments
All calculations reported in this paper were performed at TUBITAK (Turkish Scientific and Technical Research Counsel) ULAKBIM (National Academic Information Center), High Performance and Grid Computing Center (TRUBA Resources). We would like to thank TUBITAK for providing us with these excellent computational resources and service.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tekpinar, M., Yildirim, A. & Wassenaar, T.A. Molecular dynamics study of the effect of active site protonation on Helicobacter pylori 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Eur Biophys J 44, 685–696 (2015). https://doi.org/10.1007/s00249-015-1067-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-015-1067-0