Abstract
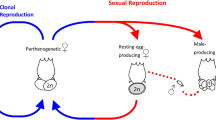
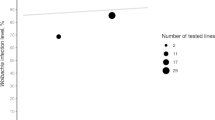
Transposable elements (TEs) are major sources of genetic variation, and mating systems are believed to play a significant role in their dynamics. For example, insertion number is expected to be higher in sexual than in asexual organisms due to the inability of TEs to colonize new genomes in the absence of sex. The goal of this study was to determine the impact of the loss of sexual reproduction on TE load. Daphnia pulex has two reproductive modes, obligate and cyclical parthenogenesis, which differ with respect to the production of diapausing eggs. Cyclical parthenogens produce them meiotically, while obligate parthenogens produce them clonally. Pokey is a TTAA-specific DNA transposon, and is a stable component of Daphnia genomes. We used a PCR-based approach, TE-Display, to estimate the number of Pokey insertions in 22 cyclic and 22 obligate isolates of D. pulex. As expected, the copy number of Pokey insertions is significantly higher in cyclic than in obligate isolates. However, the distribution of elements among isolates within each breeding system is similar, which is congruent with the recent establishment of obligate lineages from a cyclic ancestor. We also assayed 46 isolates from eight cyclic populations and found that very few Pokey insertions were observed in more than one isolate, suggesting that Pokey has been active recently. Sequencing of PCR products from the TE-Display analysis shows that Pokey inserts into both coding and noncoding regions of the genome. However, there is no obvious similarity among sequences downstream of the TTAA Pokey insertion site.





Similar content being viewed by others
References
Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein data base search program. Nucleic Acids Res 25:3389–3402
Arkhipova I, Meselson M (2000) Transposable elements in sexual and ancient asexual taxa. Proc Natl Acad Sci USA 97:14473–14477
Arkhipova I, Meselson M (2004) Deleterious transposable elements and the extinction of asexuals. Bioessays 27:76–85
Arkhipova I, Meselson M (2005) Diverse DNA transposons in rotifers of the class Bdelloidea. Proc Natl Acad Sci USA 102:11781–11786
Cavalier-Smith T (1980) How selfish is DNA? Nature 285:617–618
Crease T (1986) Mitochondrial DNA variation in Daphnia pulex Leydig producing by obligate and cyclic parthenogenesis. Ph.D. dissertation, Washington University, St. Louis, MO
Crease T, Lynch M (1991) Ribosomal DNA variation in Daphnia pulex. Mol Biol Evol 8:620–640
Crease T, Stanton D, Hebert P (1989) Polyphyletic origin of asexuality in Daphnia pulex. II. Mitochondrial DNA variation. Evolution 43:1016–1026
Doyle J, Doyle J (1987) A rapid DNA isolation procedure for small quantities of fresh leaf. Phytochem Bull 19:11–15
Eickbush T, Eickbush D (2007) Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175:477–485
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Finnegan D (1990) Transposable elements and DNA transposition in eukaryotes. Curr Opin Cell Biol 2:471–477
Gladyshev E, Meselson M, Arkhipova I (2007) A deep-branching clade of retrovirus-like retrotransposons in Bdelloid rotifers. Gene 390:136–145
Hebert P (1995) The Daphnia of North America. CD-ROM. University of Guelph, Guelph, ON
Hebert P, Beaton M, Schwartz S, Stanton D (1989) Polyphyletic origins of asexuality in Daphnia pulex: I. Breeding system variation and levels of clonal diversity. Evolution 43:1004–1015
Hebert P, Finston T (2001) Macrogeographic patterns of breeding system diversity in the Daphnia pulex group from the United States and Mexico. Heredity 87:153–161
Hebert P, Ward R, Weider L (1988) Clonal diversity patterns and breeding system variation in Daphnia pulex: an asexual–sexual complex. Evolution 42:147–159
Hebert P, Schwartz S, Ward R, Finston T (1993) Macrogeographic patterns of breeding system diversity in the Daphnia pulex group. I. Breeding systems of Canadian populations. Heredity 70:148–161
Hickey D (1982) Selfish DNA: a sexually transmitted nuclear parasite. Genetics 101:519–531
Innes D, Schwartz S, Hebert P (1986) Genotypic diversity and variation in mode of reproduction among populations in the Daphnia pulex group. Heredity 57:345–355
Kidwell M, Lisch D (2001) Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1–24
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–164
McTaggart S, Dudycha J, Omilian A, Crease T (2007) Rates of recombination in the ribosomal DNA of apomictically propagated Daphnia obtusa lines. Genetics 175:311–320
Nuzhdin S, Petrov D (2003) Transposable elements in clonal lineages: lLethal hangover from sex. Biol J Linn Soc 79:33–41
Omilian A, Taylor D (2001) Rate acceleration and long-branch attraction in a conserved gene of cryptic daphniid (Crustacea) species. Mol Biol Evol 18:2201–2212
Omilian A, Cristescu M, Dudycha J, Lynch M (2006) Ameiotic recombination in asexual lineages of Daphnia. Proc Natl Acad Sci USA 103:18638–18643
Paland S, Colbourne J, Lynch M (2005) Evolutionary history of contagious asexuality in Daphnia pulex. Evolution 59:800–813
Penton E, Crease T (2004) Evolution of the transposable element Pokey in the ribosomal DNA of the species in the subgenus Daphnia (Crustacea: Cladocera). Mol Biol Evol 21:1727–1739
Penton E, Sullender B, Crease T (2002) Pokey, a new DNA transposon in Daphnia (Cladocera: Crustacea). J Mol Evol 55:664–673
Sullender B (1993) Preliminary characterization and population genetic survey of the Daphnia rDNA transposable element, Pokey. Ph.D. dissertation, University of Oregon, Eugene, Eugene, OR
Sullender B, Crease T (2001) The behavior of a Daphnia pulex transposable element in cyclically and obligately parthenogenetic populations. J Mol Evol 53:63–69
Wright S, Schoen D (1999) Transposon dynamics and the breeding system. Genetics 107:139–148
Wright S, Le QH, Schoen D, Bureau T (2001) Population dynamics of an Ac-like transposable element in self- and cross-pollinating Arabidopsis. Genetics 158:1279–1288
Zeyl C, Bell G, Green D (1996) Sex and the spread of retrotransposon Ty3 in experimental populations of Saccharomyces cerevisiae. Genetics 143:1567–1577
Acknowledgments
Financial support for the work was provided by a research grant from the Natural Sciences and Engineering Research Council of Canada to T.J.C. We thank M. Lynch and D. Innes for generously providing some of the D. pulex isolates used in this study and A. Holliss and E. Holmes for analysis of TED fragments and sequencing of plasmid clones on the ABI3730. Comments provided by two anonymous reviewers improved an earlier version of the paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Valizadeh, P., Crease, T.J. The Association Between Breeding System and Transposable Element Dynamics in Daphnia Pulex . J Mol Evol 66, 643–654 (2008). https://doi.org/10.1007/s00239-008-9118-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9118-0