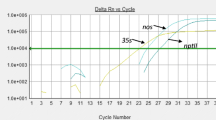
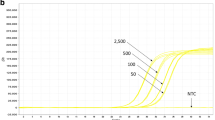
Abstract
The rapid development of many genetically modified (GM) crops in the past two decades makes it necessary to introduce an alternative strategy for routine screening and identification. In this study, we established a universal multiplex PCR detection system which will effectively reduce the number of reactions needed for sample identification. The PCR targets of this system include the six most frequently used transgenic elements: cauliflower mosaic virus (CaMV) 35S promoter, Agrobacterium tumefaciens nopaline synthase (nos) promoter, Agrobacterium tumefaciens nopaline synthase (nos) terminator, the neomycin phosphotransferase II (nptII) gene, the 5-enolpyruvylshikimate-3-phosphate synthase (CP4 epsps) gene of Agrobacterium tumefaciens strain CP4, and the phosphinothricin N-acetyltransferase (pat) gene. According to the AGBIOS database, the coverage of this detection system is 93% of commercial GM crops. This detection system could detect all certified reference materials (CRMs) at the 1.0% level. The correct combination of all the CRM amplicon patterns proved the specificity of this multiplex PCR system. Furthermore, the amplicon patterns of this multiplex PCR detection system could be used as an index of classification which will narrow the range of possible GM products. The simulation result of this multiplex PCR detection system on all commercialized 139 GM products in the AGBIOS database showed that the maximum number of PCR reactions needed to identify an unknown sample can be reduced to 13. In this study, we established a high-throughput multiplex PCR detection system with feasible sensitivity, specificity, and cost. By incorporating this detection system, the routine GM crop-detection process will meet the challenges resulting from a rapid increase in the number of GM crops in the future.




Similar content being viewed by others
Notes
The latest CRMs used in this study were ERM-BF417d (MON863 × MON810; sample no. 0046) and ERM-BF416d (MON863; sample no. 0133).
Abbreviations
- UPS:
-
Universal primer sequence
- GM:
-
Genetically modified
References
Gruere GP, Rao SR (2007) A review of international labeling policies of genetically modified food to evaluate India’s proposed rule. AgBioForum 10:51–64
Ahmed FE (2002) Detection of genetically modified organisms in foods. Trends Biotechnol 20:215–223
Miraglia M, Berdal KG, Brera C, Corbisier P, Holst-Jensen A, Kok EJ, Marvin HJ, Schimmel H, Rentsch J, van Rie JP, Zagon J (2004) Detection and traceability of genetically modified organisms in the food production chain. Food Chem Toxicol 42:1157–1180
Hubner P, Waiblinger HU, Pietsch K, Brodmann P (2001) Validation of PCR methods for quantitation of genetically modified plants in food. J AOAC Int 84:1855–1864
Holst-Jensen A (2000) Sporbarhet av genmodifiserte ingredienser i matvarer, utvikling av analyseverktøy. Genialt 4:28–33
Lipp M, Brodmann P, Pietsch K, Pauwels J, Anklam E, Borchers T, Braunschweiger G, Busch U, Eklund E, Eriksen FD, Fagan J, Fellinger A, Gaugitsch H, Hayes D, Hertel C, Hortner H, Joudrier P, Kruse L, Meyer R, Miraglia M, Muller W, Phillipp P, Popping B, Rentsch R, Wurtz A (1999) IUPAC collaborative trial study of a method to detect genetically modified soy beans and maize in dried powder. J AOAC Int 82:923–928
Trapmann S, Schimmel H, Kramer GN, Van den Eede G, Pauwels J (2002) Production of certified reference materials for the detection of genetically modified organisms. J AOAC Int 85:775–779
Hernandez M, Rodriguez-Lazaro D, Zhang D, Esteve T, Pla M, Prat S (2005) Interlaboratory transfer of a PCR multiplex method for simultaneous detection of four genetically modified maize lines: Bt11, MON810, T25, and GA21. J Agric Food Chem 53:3333–3337
Holst-Jensen A, Ronning SB, Lovseth A, Berdal KG (2003) PCR technology for screening and quantification of genetically modified organisms (GMOs). Anal Bioanal Chem 375:985–993
Onishi M, Matsuoka T, Kodama T, Kashiwaba K, Futo S, Akiyama H, Maitani T, Furui S, Oguchi T, Hino A (2005) Development of a multiplex polymerase chain reaction method for simultaneous detection of eight events of genetically modified maize. J Agric Food Chem 53:9713–9721
Xu J, Zhu S, Miao H, Huang W, Qiu M, Huang Y, Fu X, Li Y (2007) Event-specific detection of seven genetically modified soybean and maizes using multiplex-PCR coupled with oligonucleotide microarray. J Agric Food Chem 55:5575–5579
Morisset D, Dobnik D, Hamels S, Zel J, Gruden K (2008) NAIMA: target amplification strategy allowing quantitative on-chip detection of GMOs. Nucleic Acids Res 36:e118
Legendre L, Legendre P (1998) Numerical ecology. Elsevier, Amsterdam
Shuber AP, Grondin VJ, Klinger KW (1995) A simplified procedure for developing multiplex PCRs. Genome Res 5:488–493
James D, Schmidt AM, Wall E, Green M, Masri S (2003) Reliable detection and identification of genetically modified maize, soybean, and canola by multiplex PCR analysis. J Agric Food Chem 51:5829–5834
Du H, Qi G (2006) Detection of genetically modified compositions from crops in feeds. China Feed 3:33–36
Xu J, Miao H, Wu H, Huang W, Tang R, Qiu M, Wen J, Zhu S, Li Y (2006) Screening genetically modified organisms using multiplex-PCR coupled with oligonucleotide microarray. Biosens Bioelectron 22:71–77
Yang L, Guo J, Pan A, Zhang H, Zhang K, Wang Z, Zhang D (2007) Event-specific quantitative detection of nine genetically modified maizes using one novel standard reference molecule. J Agric Food Chem 55:15–24
Monsanto Technology LLC (2003) Safety assessment of Yieldgard Rootworm™ Corn. Monsanto Technology LLC, San Louis, MO, USA
Japanese Biosafety Clearing House (2004) Outline of the biological diversity risk assessment Report: Type 1 Use for MON-00863–5 x MON-00810–6 insect resistant maize. Japanese Biosafety Clearing House, Ministry of Environment, Tokyo, Japan
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, IJ., Lin, CH. & Pan, TM. Establishment of a system based on universal multiplex-PCR for screening genetically modified crops. Anal Bioanal Chem 396, 2055–2064 (2010). https://doi.org/10.1007/s00216-009-3214-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-009-3214-x