Abstract
Aims/hypothesis
In insulin-secreting cells, activation of the c-Jun NH2-terminal kinase (JNK) pathway triggers apoptosis. Whereas JNK1 and JNK2 are ubiquitously produced, JNK3 has been described exclusively in neurons. This report aims to characterise the expression and role in apoptosis of the three JNK isoforms in insulin-secreting cells exposed to cytokines.
Methods
Sections of human and mouse pancreases were used for immunohistochemistry studies with isoform-specific anti-JNK antibodies. Human, pig, mouse and rat pancreatic islets were isolated by enzymatic digestion and RNA or protein extracts were prepared. RNA and protein levels were determined by quantitative RT-PCR and western blotting respectively, using JNK-isoform-specific primers and isoform-specific antibodies; activities of the three JNK isoforms were determined by kinase assays following quantitative immunoprecipitation/depletion of JNK3. JNK silencing was performed with small interfering RNAs and apoptotic rates were determined in INS-1E cells by scoring cells displaying pycnotic nuclei.
Results
JNK3 and JNK2 mRNAs are the predominant isoforms expressed in human pancreatic islets. JNK3 is nuclear while JNK2 is also cytoplasmic. In INS-1E cells, JNK3 knockdown increases c-Jun levels and caspase-3 cleavage and sensitises cells to cytokine-induced apoptosis; in contrast, JNK1 or JNK2 knockdown is protective.
Conclusions/interpretation
In insulin-secreting cells, JNK3 plays an active role in preserving pancreatic beta cell mass from cytokine attacks. The specific localisation of JNK3 in the nucleus, its recruitment by cytokines, and its effects on key transcription factors such as c-Jun, indicate that JNK3 is certainly an important player in the transcriptional control of genes expressed in insulin-secreting cells.
Similar content being viewed by others
Introduction
Apoptosis of insulin-secreting cells is a major event leading to insulin depletion in type 1 diabetes and also contributing to islet transplantation failure [1–4]. The mechanisms leading to apoptosis have been partly deciphered and involve, among other intracellular pathways, p38 mitogen-activated protein kinase [5], nuclear factor kappa B [6], signal transducer and activator of transcription 1 [7] and activation of the stress-activated proteins c-Jun NH2-terminal kinases (JNKs) [3, 8]. Three JNK isoforms (JNK1, JNK2 and JNK3) encoded by three distinct genes have been identified [9]; JNK1 and JNK2 are ubiquitously produced, but JNK3 is essentially restricted to neurons [10, 11]. The role of JNK3 in neuronal excitotoxicity and cerebral ischaemia has been particularly well studied [12–14]; earlier reports indicated that JNK3 was the main pro-apoptotic JNK-isoform in neurons, but more recent data appear to also implicate JNK2 [15, 16].
Insulin-secreting cells and neurons share a number of genes that are predominantly or exclusively expressed in both tissues, e.g. the genes encoding: neuronal cell adhesion molecule, N-cadherin [17], neuron specific enolase [18], glutamic acid and glutamate decarboxylase [19], and islet-brain 1 (IB1) and 2 (IB2) (c-Jun-NH2-terminal kinase interacting protein [JIP] 1 and 2 respectively) [20, 21]. These characteristics probably originate from a common developmental lineage [22]. To date, functional characterisation of the JNK brain-specific isoforms in pancreatic islets has not been performed.
In this study we characterised the expression profile of the three JNK family members in human, pig, mouse and rat pancreatic islets, as well as their sub-cellular localisation and their activities following cytokine treatment. Comparative studies using small interfering RNAs (siRNAs) in rat cell lines were performed to determine the contribution of individual JNK isoforms in cytokine-mediated beta cell death.
Methods
Cell and islet culture preparations
Pancreas and islet specimens were prepared from four different deceased human multiorgan donors or from pigs that had been killed. In each case, the purity of human or pig islet preparations was >70%. The human donor’s exemption code from the Institutional Review Board is 0307M50821. Animal experimentation conformed to the Guide for Care and Use of Laboratory Animals, formulated by the National Research Council, 1996, and Swiss law on animal protection, and was authorised by the Veterinarian Office of Canton de Vaud (Lausanne, Switzerland).
Adult male rat (Wistar) pancreases were digested with collagenase solution, the fragmented tissue was washed and collected by centrifugation at 300 g and islets were separated on Ficoll density gradients. Human and pig islets were kindly provided by B. Hering (Diabetes Institute, University of Minnesota, MN, USA) and islet isolation was performed as previously described [3, 8]. Purified islets were cultured for 2 days in CMRL-1066 (Mediatech, Herndon, VA, USA) supplemented with 10% (vol./vol.) fetal bovine serum (Mediatech). The rat insulin-secreting beta cell line (INS-1E) was maintained in RPMI 1640 medium supplemented with 10% (vol./vol.) fetal bovine serum, 10 mmol/l HEPES, pH 7.4, 1 mmol/l sodium pyruvate and 50 μmol/l β-mercaptoethanol in 5% CO2 humidified atmosphere at 37°C.
Cell lysis and western blotting
Cells or fresh tissues were homogenised in cold lysis buffer as described previously [3, 8] and the protein extracts recovered by centrifugation at 20,000 g. Equal quantities of total protein lysates were resolved by SDS-PAGE and electroblotted on to polyvinylidene fluoride membranes. The blots were probed overnight with primary antibodies against: JNK1, p53 (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA); and JNK2, JNK3, c-Jun, JunB, phospho-c-Jun or cleaved caspase-3 (1:1,000; Cell Signaling Technology, Danvers, MA, USA). Equal protein loading was determined by blotting membrane against tubulin (1:5,000; Sigma-Aldrich, Buchs, Switzerland). Anti-rabbit or mouse horseradish peroxidase-conjugated secondary antibodies were used to detect protein bands with enhanced chemiluminescence reaction system (Amersham Pharmacia, Biotech Europe, Dübendorf, Switzerland).
Real-time polymerase chain reaction (RT-PCR)
RNA was extracted from tissues or cells using a commercial kit (RNAeasy Mini-kit; Qiagen, Basel, Switzerland) and 1 μg was used for reverse transcription to cDNA with Super script II enzyme (Gibco Invitrogen, Basel, Switzerland). Gene expression was measured by RT-PCR (Light-Cycler PCR System, Roche, Basel, Switzerland) as recommended by the supplier. Quantitative PCR reactions were performed in Master Mix SyBr Green (4 mmol/l; Master Mix; Eurogentec, Liege, Belgium) using 0.5 μmol/l primer mix (Electronic supplementary material [ESM] Table 1) and 2 μl RT product in a total of 20 μl.
Quantitative analysis of mRNA expression using restriction endonucleases
Degenerate primers (ESM Table 1) were designed to amplify the three human JNK mRNAs (JNK1, JNK2, JNK3 [also known as MAPK8, MAPK9 and MAPK10 respectively]) simultaneously and with high efficiency by Light-Cycler. We verified that this set of primers was equally effective in amplifying each JNK isoform by using separate plasmids coding for human JNK1, JNK2 or JNK3 genes in real-time PCRs. PCR products were purified, cloned into pGEM-t Easy Vector (Promega, Wallisellen, Switzerland) and produced by bacterial transformation. We screened 60 positive clones by PCR and determined the distinction between the three isoforms of JNK mRNA by restriction digest. Three endonuclease enzymes (PstI, EcoRV and HindIII) were used to distinguish JNK1, JNK2 and JNK3 cDNAs respectively, on agarose gels.
siRNAs design and cell transfection
Small interfering RNA duplexes against Jnk1, Jnk2, Jnk3 [23] and the green fluorescent protein (GFP) were designed (ESM Table 2) and submitted to BLAST search to ensure their specificity toward the target mRNAs. siRNAs were synthesised by Mycrosynth (Balgach, Switzerland).
Rat INS-1E cells were seeded in six-well plates and incubated overnight in antibiotic-free medium. siRNA duplexes targeting Jnk1, Jnk2 and Jnk3 were mixed with Lipofectamine2000 reagent (10 μg/ml) according to the manufacturer’s instructions (Invitrogen, Basel, Switzerland). siRNA–lipofectamine complexes were added to the cells and incubated for 2 days. Cells were exposed to a cytokine cocktail (R&D Systems, Minneapolis, MN, USA) of rat IL-1β (10 ng/ml), mouse TNFα (25 ng/ml) and rat IFNγ (150 ng/ml) for 24 h.
Immunohistochemical and immunocytochemical staining procedures
Immunohistochemistry was performed on paraffin-embedded adult human and mouse pancreatic sections (5 µm thick) collected on polylysine-coated slides. Paraffin sections were rehydrated, processed for antigen retrieval and pre-incubated with PBS containing 15% (vol./vol.) donkey serum for 1 h.
For immunocytochemistry, INS-1E cells were fixed for 15 min in paraformaldehyde 4% (wt/vol.) on ice and then pre-incubated (0.3% [vol./vol.] Triton X-100, 15% [vol./vol.] donkey serum in PBS) for 1 h. Primary antibodies were incubated overnight at 4°C after dilution in 1.5% (vol./vol.) donkey serum at the following concentrations: 1:100 rabbit anti-JNK3 (Upstate, Lake Placid, NY, USA), 1:50 mouse anti-JNK2 (Santa-Cruz Biotechnology, Santa Cruz, CA, USA) and 1:500 guinea pig anti-insulin (Dako, Carpinteria, CA, USA). After extensive washes in PBS, the sections were incubated for 2 h at room temperature in the appropriate secondary biotinylated donkey anti-mouse or donkey anti-rabbit or donkey anti-guinea pig antibody (Jackson Immunoresearch, West Grove, PA, USA) diluted 1:500 in PBS. The sections were thoroughly rinsed in PBS and incubated for 2 h at room temperature with streptavidin–biotin–peroxidase complex (Vectastain ABC kit, Vector Laboratories, Burlingame, CA, USA). Immunolabelling was revealed after three washes in PBS using 2,3′ diaminobenzidene as substrate diluted 1:10 in buffer according to the manufacturer’s instructions (Roche). Sections were then dehydrated and mounted in Eukitt (O. Kindler, Freiburg, Germany). The specificity of the immunostaining was confirmed by omission of the first antibody or use of preadsorption of primary antibodies with the immunising peptide. Images were acquired by optical microscopy.
Immunoprecipitation and JNK kinase assays
Pre-cleared protein extracts (300 μg) were mixed overnight at 4°C with 1 μg of anti-JNK3 antibody. Protein-G agarose beads (50 μl; Calbiochem, San Diego, CA, USA) were then added for an additional incubation of 4 h. Immobilised protein–antibody complexes were washed three times in cold PBS, collected by centrifugation at 2,000 g and lysed in kinase buffer (20 mmol/l HEPES pH 7.5, 20 mmol/l β-glycerophosphate, 10 mmol/l MgCl2). Supernatant fractions were controlled for potentially remaining JNK3 by western blotting. Isoform-specific JNK activities were determined by in vitro reactions using glutathione S-transferase (GST)–c-Jun (amino acids 1–79) beads mixed to [γ-33P]ATP (Amersham Biosciences, Little Chalfont, UK). The kinase reactions were performed for 30 min at 30°C and stopped by adding SDS sample buffer (100 mmol/l Tris, pH 6.8, 2% [wt/vol.] SDS, 10% [vol./vol.] glycerol, 5% [vol./vol.] 2-β-mercaptoethanol, 0.25% [wt/vol.] bromophenol blue). Phosphorylation of substrate proteins was detected by autoradiography of dried gels (BioMax MS films; Kodak, GE Healthcare, Amersham Biosciences, Little Chalfont, UK).
Apoptosis assay
Apoptosis was determined by scoring cells displaying pycnotic nuclei stained with the DNA-binding dyes Hoechst 33342 (10 μg/ml; Sigma-Aldrich, St Louis, MO, USA) and propidium iodide (5 μg/ml; Sigma-Aldrich, St Louis, MO, USA), as described previously [24], and visualised under an inverted fluorescence microscope.
Statistics
All experiments reported were performed a minimum of three times (i.e. n = 3–9 for all data). Data are given as means ± SEM. Statistical analysis of multiple comparisons was determined by one-way ANOVA (post hoc comparisons, Scheffe’s test). The differences found between the experimental groups were considered statistically significant at p < 0.05 or p < 0.01.
Results
JNK3 is highly produced in pancreatic islets
To determine in situ levels of the JNK3 protein in pancreatic islets, we performed immunohistochemistry studies on human and mouse pancreas sections using specific antibodies against JNK3 and insulin. Human (Fig. 1a) and mouse (Fig. 1a) pancreas sections were stained with haematoxylin and eosin. JNK3-positive cells (Fig. 1a) were also positive for insulin (Fig. 1a). While insulin staining was mostly cytoplasmic, JNK3 was nuclear. The specificity of the JNK3 antibody was ascertained by western blotting using whole-brain protein extracts originating from Jnk1, -2 or -3 knockout mice (Fig. 1b).
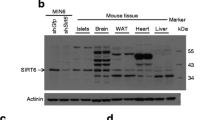
Analyses of protein production, with a immunohistochemistry analysis using haematoxylin and eosin staining (H&E) of pancreatic tissue sections as shown, JNK3 immuno-staining of representative pancreatic islet cells (arrows), insulin immuno-staining (INS) of representative pancreatic islet cells and negative controls (CTL). Original magnification, ×20; scale bar, 50 μm. b The specificity of the antibody was tested by western blotting using whole-brain protein extracts derived from wild-type (WT) and Jnk1, Jnk2 or Jnk3 knockout mice. c–e Western blot analysis of protein extracts prepared from different species and used in experiments with monoclonal anti-JNK3 and anti-tubulin antibodies, i.e. in c whole pancreas and isolated islets from human or pig, d rat tissues and e different insulin-secreting (INS-1E, βTC3, MIN6) and non-secreting (HaCaT, HEK293T) cell lines. Protein extracts from mouse and rat whole brain were used e as positive controls for JNK3 detection. Data are representative of three independent experiments (n = 3)
We next examined by western blot analysis the levels of JNK3 in different tissues. The blots revealed that JNK3 occurs in high levels in human and pig islets, but was not detected in either of the whole-pancreas species (Fig. 1c). In the rat, JNK3 production was restricted to the brain and pancreatic islets (Fig. 1d), whereas JNK1 and JNK2 were ubiquitously produced at similar levels in the tissues tested (data not shown).
In insulin-producing cell lines, production of JNK3 was restricted to the rat beta cell line INS-1E; JNK3 was not detected in mouse βTC3 and MIN6 cell lines (Fig. 1e). Antibody specificity was confirmed using protein extracts from human keratinocyte (HaCaT) and human embryonic kidney (HEK)293T cell lines, which do not produce JNK3; protein extracts from mouse or rat brain were used as positive controls for JNK3 detection (Fig. 1e).
Quantitative analysis of JNK1, JNK2 and JNK3 mRNA levels in pancreatic islets
To determine the endogenous levels of JNK1, JNK2 and JNK3 mRNA in different tissues, quantitative real-time PCR (Light-Cycler) was performed with primers specific for each JNK isoform (Fig. 2a). In human purified islets, JNK3 mRNA was expressed at levels similar to those found in the brain. A strong reduction in JNK3 mRNA expression was observed after 2 days of culture (Fig. 2b). No amplification product was obtained in these conditions from the other tissues tested and in particular from whole pancreas. JNK1 and JNK2 transcripts were detected in all examined human tissues with predominant expression of JNK1 mRNA in the brain and lower levels in pancreatic islets (Fig. 2b). No major alteration of JNK1 or JNK2 mRNA levels was observed in purified vs cultured islets. Similar observations were noted for rat and mouse tissues (Fig. 2c).
mRNA expression analyses. a Isoform-specific primers were tested by PCR using plasmids (1 ng) encoding JNK1, JNK2 and JNK3 cDNAs. The resulting PCR products were resolved on 1% agarose gels containing ethidium bromide and visualised by ultra-violet fluorescence. b Total RNA was isolated from human and c murine tissues, and reverse-transcribed to cDNA before PCR amplification (Light-Cycler). The sample RNAs were normalised to tubulin (Tuba1a) mRNA (internal control); the relative levels of the mRNA for each JNK gene were set to 100% in the brain samples of each species. ‘Purified’ or ‘cultured’ indicates human islets immediately after the isolation process or 2 days after culture. White bars, JNK1; grey bars, JNK2; black bars, JNK3 (b). c Black bars, rat; white bars, mouse. Data are the mean of n = 4 (b) or n = 3 (c) independent experiments. d Relative amounts of JNK isoforms in human pancreatic islets. Degenerate primers were used to simultaneously amplify the genes encoding all three JNK isoforms. PCR products were then cloned into the pGEM-T vector. Sixty clones were screened by PCR and the identity of each JNK gene was determined by enzymatic digestion. PstI, EcoRV and HindIII were used to distinguish JNK1, JNK2 and JNK3, respectively. DNA fragments were analysed on agarose gels stained with ethidium bromide. Data are representative of three independent experiments
To determine the relative levels of JNK1, JNK2 and JNK3 mRNA in isolated human islets, we used degenerate primers to amplify all three mRNAs simultaneously. We confirmed that this set of primers was equally effective in amplifying each JNK gene separately (data not shown). PCR products were then cloned and analysed by enzymatic digestion to allow discrimination of the respective JNK isoforms. JNK2 and JNK3 isoforms (EcoRV and HindIII fragments, respectively) represented 45% each of the total JNK clones analysed, while 10% of the clones were of JNK1 origin (Fig. 2d). Thus JNK2 and JNK3 represent the two most highly expressed JNK isoforms in human pancreatic islets.
JNK3 is a component of a functional signalling pathway activated by cytokines
To address whether JNK3 is activated after exposure to cytokines, INS-1E cells were treated with IL-1β, TNFα and IFNγ individually or in combination (Cyto-Mix) for 1 h. JNK3 was then quantitatively immuno-precipitated from the protein extracts; we verified by western blotting that following JNK3 immuno-precipitation, JNK3 was absent from the supernatant fractions and that no JNK1/2 could be detected in the pellets (not shown). JNK1/2 (supernatant fractions) vs JNK3 (pellets) activity was assessed by in vitro kinase assays using GST-c-Jun as a substrate. All three isoforms of JNK were potently activated by IL-1β and to a lesser extent by TNFα; in particular, JNK3 appeared to be activated to a similar extent to JNK1 and JNK2 together (Fig. 3). This indicates that JNK3 can be potently activated by cytokines in beta cells.
Kinase activity analyses. INS-1E cells were exposed to the cytokines IL-1β, TNFα, or IFNγ individually or in combination (Cyto-Mix) for 1 h and protein extracts were then prepared. Pre-cleared extracts (300 μg) were used for JNK3 immuno-precipitation with protein A-agarose beads. The supernatant fractions (p-JNK1/2) or the immuno-complexes (p-JNK3) were tested in in vitro kinase reactions with GST–c-Jun as a substrate and [γ-33P]ATP. Part of the supernatant fractions was used for western blot analysis to ascertain equal loading using the anti-JNK1/2 antibody. Data shown are the mean of three independent experiments
JNK3 is nuclear whereas JNK2 is both cytoplasmic and nuclear
Immunocytochemistry studies were carried out to determine the sub-cellular localisation of JNK3 and JNK2 in INS-1E cells. As shown in Fig. 4a, JNK2 was detected in both cytoplasmic and nuclear compartments, while JNK3 was restricted to the nucleus (see also Fig. 1a). The specificity of the JNK3 (Fig. 1b) and JNK2 (Fig. 4b) antibodies was ascertained by western blotting using whole-brain protein extracts originating from Jnk1, -2 or -3 knockout mice respectively.
a Subcellular localisation analyses in INS-1E cells. Immunocytochemical staining localised JNK3 in the nucleus exclusively, while JNK2 staining appeared in the cytoplasm, with a nuclear staining component that cannot be excluded. Negative controls (CTL) used non-immunised rabbit serum. Original magnification: ×10 and ×20 on left and right, respectively; scale bar, 50 μm. b Specificity of the JNK2 antibody was tested by western blotting using whole-brain protein extracts derived from Jnk1, Jnk2 or Jnk3 knockout mice. WT, wild-type
Knockdown of JNK3 in INS-1E cells results in higher c-Jun levels
The different intracellular (nuclear vs nuclear/cytoplasmic) localisations of JNK3 and JNK2 in INS-1E cells suggest that these two isoforms might differentially control the cellular responses elicited by the same input stresses. To investigate whether the individual JNK isoforms might differentially transduce cytokine signalling, INS-1 cells were transfected with siRNAs specific for the JNK3, JNK2 and JNK1 isoforms, while siRNA against GFP (siGFP) was used as a negative control. JNK3 knockdown decreased JNK3 protein levels up to 90% with little if any effect on JNK1 or JNK2 (Fig. 5a). JNK1 and JNK2 knockdown were also performed and showed a specific reduction of JNK1 (85%) and JNK2 (90%), respectively, with little if any effect on the other JNK isoforms (data not shown and Fig. 6a).
a Transfection efficiency of siRNAs. INS-1E cells were transiently transfected with Jnk3 siRNA (si3) at different doses as shown using lipofectamine 2000. siGFP (120 pmol) was used as a control. JNK3 protein knockdown was tested by western blotting using an anti-JNK3 antibody; anti-JNK1 and anti-JNK2 antibodies were used to assess the specificity of the assay. Equal protein loading was determined with a tubulin antibody. b, c Transfected cells (si3, si2 and si1) were treated with or without cytokines (Cyto-Mix) for 1 h and extracted proteins were resolved by western blotting to determine c-Jun (b) and JunB or phospho-c-Jun (P-c-Jun) (c) levels. Equal protein loading was verified with an anti-tubulin antibody. Optical density for each protein band was quantified by densitometric scanning; ratios normalised to tubulin (controls set to 1) are indicated. Data are representative of n = 4 (a) or n = 3 (b, c) independent experiments
Effects of JNK1, JNK2 and JNK3 knockdown on cytokine-mediated apoptosis. a INS-1E cells were transfected with the siRNAs described above, i.e. Jnk1 (si1), Jnk2 (si2) and Jnk3 (si3) or siGFP, for 48 h. Transfected cells were then treated for 24 h with or without the cytokine cocktail (Cyto-Mix). JNK1, JNK2 and JNK3 protein knockdowns were tested by western blot using specific antibodies and protein normalisation (tubulin) as above (Fig. 5a). Cell apoptosis was determined by scoring pycnotic nuclei visualised by Hoechst and propidium iodide dyes under a fluorescence microscope. *p < 0.05 or **p < 0.01 for Mix-si3/si3II vs Mix-siGFP, for Mix-si2 vs Mix-siGFP and for Mix-si1 vs Mix-siGFP. White bars, basal; grey bars, siRNA; black bars, siRNA-cytokines. b Whole cell lysates were analysed by western blotting for cleaved caspase-3 (19 kDa fragment) and p53 expression. Optical density for each protein band was quantified by densitometric scanning; ratios normalised to tubulin (controls set to 1) are indicated. Data are representative of n = 3 independent experiments
Western blot experiments indicate that JNK3 knockdown increased the protein level of c-Jun; cytokines had a similar, but more pronounced, effect on c-Jun induction (Fig. 5b). Combined JNK3 knockdown and cytokines resulted in an even higher phosphorylation of c-Jun than in each of the separate conditions (Fig. 5c). In contrast, JNK2 knockdown had no detectable effect on levels of c-Jun production in the basal state (Fig. 5b). Both, JNK1 or JNK2 deficiency decreased c-Jun phosphorylation induced by cytokines (Fig. 5c).
Previous studies have shown that the activator protein-1 component JunB is upregulated by cytokines [25, 26]. Cytokines increased JunB levels as expected; JNK1 and to a lesser extent JNK3, but not JNK2 knockdown appeared to decrease cytokine-induced JunB upregulation (Fig. 5c).
JNK3 knockdown increases apoptosis
To determine the role of the different JNK isoforms in cytokine-induced apoptosis, INS-1E cells were transfected with the siRNAs targeting the different JNK genes and were exposed for 24 h to the cytokine cocktail. JNK2 or JNK1 knockdown significantly protected beta cells against cytokine-induced apoptosis and improved beta cell viability by 60 and 40% respectively compared with control siGFP-transfected cells (Fig. 6a). In contrast, JNK3 knockdown strongly enhanced the effect of cytokines on apoptosis. As control, similar results were obtained when using a different siRNA (si3II) against Jnk3 [23] (Fig. 6a). The pro-apoptotic effect of JNK3 knockdown was associated with an increase in caspase-3 cleavage (as evidenced by the appearance of a 19 kDa fragment) at basal or under stress conditions (Cyto-Mix) (Fig. 6b). A similar pattern of cleaved caspace-3 was observed with the second siRNA (si3II) against Jnk3 (data not shown). In contrast, JNK1 or JNK2 knockdown were able to block caspase-3 activation. None of these conditions appeared to affect p53 stability (Fig. 6b).
These data were further sustained by transfecting the siRNAS in βTC-3 cells. Because these cells lack JNK3 (see Fig. 1e), JNK3 knockdown was not expected to have any effect on viability of these cells. Indeed, JNK3 knockdown did not increase apoptosis compared with the siGFP control under cytokine treatments (data not shown).
Discussion
We describe here the content of JNK3 in pancreatic islets, where it is found at levels similar to those measured in the brain. Our data indicate a protective role of JNK3 against cytokine-induced beta cell death. Whereas knockdown of JNK1 or JNK2 is anti-apoptotic, JNK3 knockdown potentiates cell death in INS-1E cells. Both effects correlate well with the levels of and phosphorylation of c-Jun, which are elevated in the absence of JNK3 and reduced in the absence of the JNK1 or JNK2 isoforms.
The phenotype of JNK3 knockdown is similar to that observed with JunB knockdown, which also increases apoptosis [26]. Interestingly, we observed a JunB decrease in cells lacking JNK3. The question of whether JunB is a major determinant of apoptosis in JNK3-deficient insulin-secreting cells undoubtedly warrants further investigation. However, the situation is further complicated in JNK1 or JNK2 knockdown, where increased levels of JunB correlates with protection (JNK2), but not with JNK1 (decreased apoptosis and decreased JunB levels).
In other cell types including fibroblasts, it has been shown that inactive JNK targets c-Jun to ubiquitination, whereas activation of JNK improves c-Jun stability [27, 28]. Conflicting studies, however, have shown that JNK-mediated phosphorylation of c-Jun accelerates its ubiquitination in T cells [29, 30]. In fibroblasts and in neurons, JNK is usually observed essentially in the cytoplasm, with a fraction of it translocating to the nucleus following activation [31, 32]. We observed very pronounced differences in the intracellular distribution of JNK2 (nuclear and cytoplasmic) vs JNK3, with JNK3 being essentially nuclear. Thus, insulin-secreting cells are clearly distinct from other cell types in that they have high amounts of JNK (JNK3) in the nucleus at basal states.
It is not clear whether the main differences between JNK3 and JNK2 or JNK1 knockdown that we observed are related to the levels of each kinase as such or potentially to the specific intracellular localisation of JNK3 vs JNK2. However, the importance of the intracellular localisation of the different JNK isoforms towards apoptosis has already been addressed [32–34]: in neurons, blockage of nuclear JNKs appears neuroprotective, whereas blocking cytoplasmic JNKs has a strong pro-apoptotic activity [34, 35]. It should, however, be noted that in these experiments, ‘cytoplasmic’ blockade of JNK resulted in depletion from the nucleus of JNKs, which bind to the cytosolic IB1/JIP-1 protein that is exogenously expressed in the cytoplasm (J. Puyal, Department of Cellular Biology and Morphology, University of Lausanne, Lausanne, Switzerland, personal communication). A similar observation had already been made by Dickens, who showed that expression of IB1/JIP-1 ‘traps’ JNK in the cytoplasm, depleting the nuclear compartment [35]. Thus, these and our data indicate that depletion of nuclear JNK is toxic, therefore suggesting that nuclear JNK has a protective role in neurons and insulin-secreting cells.
The effects of JNK3 on apoptosis might be linked to the control of key factors directly contributing to the apoptotic machinery and controlled by c-Jun, such as the Fas-ligand [36, 37] or the gene encoding p53 [38]; we did not detect any effect of JNK3 knockdown on p53 expression in our conditions. However, the effect might also be indirect. Indeed, the JNK–c-Jun signalling pathway is known to control expression of the insulin promoter by repressing cAMP-induced insulin transcriptional activity [39]. Moreover, pancreatic and duodenal homeobox 1 (PDX-1), an important activator of the insulin promoter, is translocated from the nucleus to the cytoplasm of pancreatic beta cells in response to oxidative stress [40, 41]. Dominant negative forms of JNK inhibit oxidative stress-induced PDX-1 translocation, suggestive of another way by which JNK, and in particular JNK3, might regulate activity of the insulin promoter. Given the known protective role of insulin acting in an autocrine manner on insulin-secreting cells, JNK3 might potentially control apoptosis through the regulation of insulin expression. Indeed, JNK3 knockdown may affect insulin expression (data not shown).
Pancreatic beta cells and neurons share many phenotypic traits such as the expression of neurotrophin receptors [42] or neuron-specific factors [17–21]. These similarities may be partly explained by the action of a key transcriptional repressor known as ‘transcriptional repressor element 1 silencing transcription factor’ (REST/NRSF). REST acts as a transcriptional repressor that controls the transcription of neuronal genes in non-neuronal tissues, but is not produced in neurons and endocrine pancreas; accordingly, most of its target genes are highly expressed in these two cell types, because they lack REST expression [43–45]. Genes targeted by REST encode neuronal genes expressed in beta cells, such as connexin 36, synaptophysin, secretogranin II and neuronal receptors like glutamate receptor 2 and brain-derived neurotrophin factor [45], as well as IB1 [20] or IB2 [21] transcription factors and adhesion molecules [17]. The regulatory region of the JNK3 gene contains a potentially functional repressor element-1 that might interact with REST (http://bioinformatics.leeds.ac.uk/group/online/RE1db/re1db_home.htm, accessed 21 May 2009) [46]. These observations are fully consistent with the high level of JNK3 that we found in pancreatic islets.
Altogether, JNK1 or JNK2 vs JNK3 deficiencies exert opposite effects on apoptosis in insulin-secreting cells. The specific localisation of JNK3 in the nucleus, its recruitment by cytokines and its role in downregulating c-Jun levels suggest that JNK3 may play an important role in the transcriptional control of genes expressed in insulin-secreting cells.
Abbreviations
- GFP:
-
Green fluorescent protein
- GST:
-
Glutathione S-transferase
- HaCaT:
-
Human keratinocyte
- HEK:
-
Human embryonic kidney
- IB1:
-
Islet-brain 1
- IB2:
-
Islet-brain 2
- JIP:
-
c-Jun-NH2-terminal kinase interacting protein
- JNK:
-
c-Jun-NH2-terminal kinase
- PDX-1:
-
Pancreatic and duodenal homeobox 1
- REST:
-
Transcriptional repressor element 1 silencing transcription factor
- siRNA:
-
Small interfering RNA
References
Mandrup-Poulsen T (1996) The role of interleukin-1 in the pathogenesis of IDDM. Diabetologia 39:1005–1029
Helqvist S, Zumsteg UW, Spinas GA et al (1991) Repetitive exposure of pancreatic islets to interleukin-1 beta. An in vitro model of pre-diabetes? Autoimmunity 10:311–318
Abdelli S, Ansite J, Roduit R et al (2004) Intracellular stress signaling pathways activated during human islet preparation and following acute cytokine exposure. Diabetes 53:2815–2823
Barshes NR, Wyllie S, Goss JA (2005) Inflammation-mediated dysfunction and apoptosis in pancreatic islet transplantation: implications for intrahepatic grafts. J Leukoc Biol 77:587–597
Saldeen J, Lee JC, Welsh N (2001) Role of p38 mitogen-activated protein kinase (p38 MAPK) in cytokine-induced rat islet cell apoptosis. Biochem Pharmacol 61:1561–1569
Ortis F, Pirot P, Naamane N et al (2008) Induction of nuclear factor-kappaB and its downstream genes by TNF-alpha and IL-1beta has a pro-apoptotic role in pancreatic beta cells. Diabetologia 51:1213–1225
Gysemans CA, Ladrière L, Callewaert H et al (2005) Disruption of the gamma-interferon signaling pathway at the level of signal transducer and activator of transcription-1 prevents immune destruction of beta-cells. Diabetes 54:2396–2403
Abdelli S, Abderrahmani A, Hering BJ, Beckmann JS, Bonny C (2007) The c-Jun N-terminal kinase JNK participates in cytokine- and isolation stress-induced rat pancreatic islet apoptosis. Diabetologia 50:1660–1669
Gupta S, Barrett T, Whitmarsh AJ et al (1996) Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J 15:2760–2770
Ip YT, Davis RJ (1998) Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol 10:205–219
Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103:239–252
Yang DD, Kuan CY, Whitmarsh AJ et al (1997) Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature 389:865–870
Tian H, Zhang G, Li H, Zhang Q (2003) Antioxidant NAC and AMPA/KA receptor antagonist DNQX inhibited JNK3 activation following global ischemia in rat hippocampus. Neurosci Res 46:191–197
Brecht S, Kirchhof R, Chromik A et al (2005) Specific pathophysiological functions of JNK isoforms in the brain. Eur J Neurosci 21:363–377
Coffey ET, Smiciene G, Hongisto V et al (2002) c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J Neurosci 22:4335–4345
Ries V, Silva RM, Oo TF et al (2008) JNK2 and JNK3 combined are essential for apoptosis in dopamine neurons of the substantia nigra, but are not required for axon degeneration. J Neurochem 107:1578–1588
Møller CJ, Christgau S, Williamson MR et al (1992) Differential expression of neural cell adhesion molecule and cadherins in pancreatic islets, glucagonomas, and insulinomas. Mol Endocrinol 6:1332–1342
Bishop AE, Polak JM, Facer P, Ferri GL, Marangos PJ, Pearse AG (1982) Neuron specific enolase: a common marker for the endocrine cells and innervation of the gut and pancreas. Gastroenterology 83:902–915
Okada Y, Taniguchi H, Schimada C (1976) High concentration of GABA and high glutamate decarboxylase activity in rat pancreatic islets and human insulinoma. Science 194:620–622
Bonny C, Nicod P, Waeber G (1998) IB1, a JIP-1-related nuclear protein present in insulin-secreting cells. J Biol Chem 273:1843–1846
Negri S, Oberson A, Steinmann M et al (2000) cDNA cloning and mapping of a novel islet-brain/JNK-interacting protein. Genomics 64:324–330
Dubois PM (1989) Ontogeny of the endocrine pancreas. Horm Res 32:53–60
Wang Y, Luo W, Reiser G (2007) Proteinase-activated receptor-1 and -2 induce the release of chemokine GRO/CINC-1 from rat astrocytes via differential activation of JNK isoforms, evoking multiple protective pathways in brain. Biochem J 401:65–78
Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF (2001) Cell-permeable peptide inhibitors of JNK: novel blockers of beta-cell death. Diabetes 50:77–82
Kutlu B, Cardozo AK, Darville MI et al (2003) Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 52:2701–2719
Gurzov EN, Ortis F, Bakiri L, Wagner EF, Eizirik DL (2008) JunB inhibits ER stress and apoptosis in pancreatic beta cells. Plos ONE 3:e3030
Fuchs SY, Dolan L, Davis RJ, Ronai Z (1996) Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13:1531–1535
Fuchs SY, Xie B, Adler V, Fried VA, Davis RJ, Ronai Z (1997) c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J Biol Chem 272:32163–32168
Nateri AS, Riera-Sans L, Da Costa C, Behrens A (2004) The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303:1374–1378
Gao M, Labuda T, Xia Y et al (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306:271–275
Lee JK, Park J, Lee YD, Lee SH, Han PL (1999) Distinct localization of SAPK isoforms in neurons of adult mouse brain implies multiple signaling modes of SAPK pathway. Brain Res Mol Brain Res 70:116–124
Björkblom B, Vainio JC, Hongisto V, Herdegen T, Courtney MJ, Coffey ET (2008) All JNKs can kill, but nuclear localization is critical for neuronal death. J Biol Chem 283:19704–19713
Coffey ET, Hongisto V, Dickens M, Davis RJ, Courtney MJ (2000) Dual roles for c-Jun N-terminal kinase in developmental and stress responses in cerebellar granule neurons. J Neurosci 20:7602–7613
Harding TC, Xue L, Bienemann A et al (2001) Inhibition of JNK by overexpression of the JNL binding domain of JIP-1 prevents apoptosis in sympathetic neurons. J Biol Chem 276:4531–4534
Dickens M, Rogers JS, Cavanagh J et al (1997) A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277:693–696
Le-Niculescu H, Bonfoco E, Kasuya Y, Claret FX, Green DR, Karin M (1999) Withdrawal of survival factors results in activation of the JNK pathway in neuronal cells leading to Fas ligand induction and cell death. Mol Cell Biol 19:751–763
Kolbus A, Herr I, Schreiber M, Debatin KM, Wagner EF, Angel P (2000) c-Jun-dependent CD95-L expression is a rate-limiting step in the induction of apoptosis by alkylating agents. Mol Cell Biol 20:575–582
Shaulian E, Schreiber M, Piu F, Beeche M, Wagner EF, Karin M (2000) The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell 103:897–907
Inagaki N, Maekawa T, Sudo T, Ishii S, Seino Y, Imura H (1992) c-Jun represses the human insulin promoter activity that depends on multiple cAMP response elements. Proc Natl Acad Sci U S A 89:1045–1049
Kawamori D, Kajimoto Y, Kaneto H et al (2003) Oxidative stress induces nucleo-cytoplasmic translocation of pancreatic transcription factor PDX-1 through activation of c-Jun NH(2)-terminal kinase. Diabetes 52:2896–2904
Kawamori D, Kaneto H, Nakatani Y et al (2006) The forkhead transcription factor Foxo1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J Biol Chem 281:1091–1098
Kanaka-Gantenbein C, Dicou E, Czernichow P, Scharfmann R (1995) Presence of nerve growth factor and its receptors in an in vitro model of islet cell development: implication in normal islet morphogenesis. Endocrinology 36:3154–3162
Chong JA, Tapia-Ramírez J, Kim S et al (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80:949–957
Atouf F, Czernichow P, Scharfmann R (1997) Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. J Biol Chem 272:1929–1934
Hohl M, Thiel G (2005) Cell type-specific regulation of RE-1 silencing transcription factor (REST) target genes. Eur J Neurosci 22:2216–2230
Bruce AW, Donaldson IJ, Wood IC et al (2004) Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A 101:10458–10463
Acknowledgements
We thank F. Maurer for always helpful discussions and a critical reading of the manuscript. This study was supported by grants from the Botnar Foundation and the Swiss National Science Foundation (FNS 320000-118193).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
Sequences of the oligonucleotides used for the Light-Cycler (PDF 19 kb)
ESM Table 2
Sequence for the JNKs and scrambled siRNAs used for transfection studies (PDF 13 kb)
Rights and permissions
About this article
Cite this article
Abdelli, S., Puyal, J., Bielmann, C. et al. JNK3 is abundant in insulin-secreting cells and protects against cytokine-induced apoptosis. Diabetologia 52, 1871–1880 (2009). https://doi.org/10.1007/s00125-009-1431-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-009-1431-7