Abstract
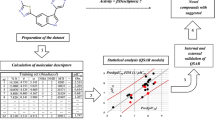
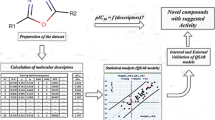
Quantitative structure–activity relationship (QSAR) studies were performed on a series of 21 thiazolidine-2,4-dione derivatives to find the structural requirements for PIM-2 kinase inhibitory activity by two-dimensional (2D-QSAR), group-based (G-QSAR) and three-dimensional (3D-QSAR) studies. In the present study, widely used technique viz. stepwise forward–backward (SW-FB) has been applied for the development of 2D- and G-QSAR as variable selection method. The statistically significant best 2D-QSAR model was developed by partial least squares regression (PLSR) having r 2 = 0.78, q 2 = 0.63 with pred_r 2 = 0.78. The statistically significant best G-QSAR model was developed by PLSR method having r 2 = 0.89, q 2 = 0.79 and pred_r 2 = 0.82. The 3D-QSAR studies were performed by k-nearest neighbor molecular field analysis along with genetic algorithm method which showed q 2 = 0.64 and pred_r 2 = 0.94. A docking study revealed the binding orientations of these inhibitors at active site amino acid residues (PHE 43, ASP 124, ASP 182 and GLU 83) of PIM-2 enzyme (PDB ID: 3IWI). The results of this study may be useful to (medicinal) chemists to design more potent thiazolidine-2,4-dione analogs as PIM-2 kinase inhibitors.









Similar content being viewed by others
References
Ajmani S, Jadhav K, Kulkarni SA (2006) Three-dimensional QSAR using the k-nearest neighbor method and its interpretation. J Chem Inf Model 46:24–31
Ajmani S, Jadhav K, Kulkarni SA (2009) Group-based QSAR (GQSAR): mitigating interpretation challenges in QSAR. QSAR Comb Sci 28:36–41
Ajmani S, Jadhav K, Kulkarni SA (2010) A comprehensive structure–activity analysis of protein kinase B-alpha (Akt1) inhibitors. J Mol Graph Model 28:683–694
Asati V, Mahapatra DK, Bharti SK (2014) Thiazolidine-2,4-diones as multi-targeted scaffold in medicinal chemistry: potential anticancer agents. Eur J Med Chem 87:814–833
Baumann K (2002) An alignment-independent versatile structure descriptor for QSAR and QSPR based on the distribution of molecular features. J Chem Inf Comput Sci 42:26–35
Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J (2010) PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica 95:1004–1015
Chang MW, Ayeni C, Breuer S, Torbett BE (2010) Virtual screening for HIV protease inhibitors: a comparison of AutoDock 4 and Vina. PLoS One 5(8):e11955
Dakin LA, Block MH, Chen H, Code E, Dowling JE, Feng X, Ferguson AD, Green I, Hird AW, Howard T, Keeton EK, Lamb ML, Lyne PD, Pollard H, Read J, Wu AJ, Zhang T, Zheng X (2012) Discovery of novel benzylidene-1,3-thiazolidine-2,4-diones as potent and selective inhibitors of the PIM-1, PIM-2, and PIM-3 protein kinases. Bioorg Med Chem Lett 22:4599–4604
Forshell LP, Li Y, Forshell TZ, Rudelius M, Nilsson L, Keller U, Nilsson J (2011) The direct Myc target PIM-3 cooperates with other PIM kinases in supporting viability of Myc-induced B-cell lymphomas. Oncotarget 2:448–460
Ghosh P, Bagchi MC (2009) QSAR modeling for quinoxaline derivatives using genetic algorithm and simulated annealing based feature selection. Curr Med Chem 16:4032–4048
Golbraikh A, Tropsha A (2002) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput Aided Mol Des 16:357–369
Halgren TA (1996) Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. J Comput Chem 17:553–586
Hasegawa K, Kimura T, Funatsu K (1999) GA strategy for variable selection in QSAR studies: enhancement of comparative molecular binding energy analysis by GA-based PLS method. Quant Struct Act Relatsh 18:262–272
http://vina.scripps.edu/ (Accessed on 2 Sep 2014)
Lee J, Park J, Hong VS (2014) Synthesis and evaluation of 5-(3-(pyrazin-2-yl)benzylidene)thiazolidine-2,4-dione derivatives as pan–PIM kinases inhibitors. Chem Pharm Bull 62(9):906–914
Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A (2004) Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol 24:6104–6115
Möröy T, Verbeek S, Ma A, Achacoso P, Berns A, Alt F (1991) E mu N and E mu L-myc cooperate with E mu pim-1 to generate lymphoid tumors at high frequency in double-transgenic mice. Oncogene 6:1941–1948
Nawijn MC, Alendar A, Berns A (2011) For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11:23–34
Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS (2002) Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in autodock. Proteins Struct Funct Genet 46:34–40
Sahu NK, Shahi S, Sharma MC, Kohli DV (2011) QSAR studies on imidazopyridazine derivatives as PfPK7 inhibitors. Mol Simul 37:752–765
Seeliger D, Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24:417–422
Sharaf MA, Illman DL, Kowalski BR (1986) Chemometrics. Wiley, New York
Shen M, Xiao Y, Golbraikh A, Gombar VK, Tropsha A (2003) Development and validation of k-nearest-neighbor QSPR models of metabolic stability of drug candidates. J Med Chem 46:3013–3020
Stigliani JL, Bernardes-Génisson V, Bernadoua J, Pratviela G (2012) Cross-docking study on InhA inhibitors: a combination of Autodock Vina and PM6-DH2 simulations to retrieve bio-active conformations. Org Biomol Chem 10:6341–6345
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comp Chem 31:455–461
Van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, Berns A (1989) Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56:673–682
VLife Sciences Technology Pvt. Ltd., Pune. www.vlifesciences.com
Wold S (1995) PLS for multivariate linear modelling. In: van de Waterbeemd H (ed) QSAR: chemometric methods in molecular design. Wiley-VCH, Weinheim, pp 195–218
Zhang Y, Wang Z, Li X, Magnuson NS (2008) Pim kinase-dependent inhibition of c-Myc degradation. Oncogene 27:4809–4819
Acknowledgments
The authors gratefully acknowledge Indian Council of Medical Research (ICMR), New Delhi, for providing Senior Research Fellowship (SRF) to Mr. Vivek Asati (SRF No. 45/28/2013-PHA/BMS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Asati, V., Bharti, S.K. QSAR studies for some thiazolidine-2,4-dione derivatives as PIM-2 kinase inhibitors. Med Chem Res 25, 1329–1339 (2016). https://doi.org/10.1007/s00044-016-1577-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-016-1577-z