Abstract
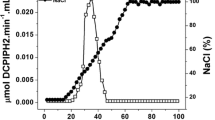
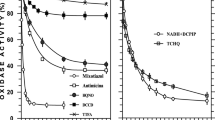
Iminodiacetate (IDA) is a xenobiotic intermediate common to both aerobic and anaerobic metabolism of nitrilotriacetate (NTA). It is formed by either NTA monooxygenase or NTA dehydrogenase. In this paper the detection and characterization of a membrane-bound iminodiacete dehydrogenase (IDA-DH) from Chelatobacter heintzii ATCC 29600 is reported, which oxidizes IDA to glycine and glyoxylate. Out of 15 compounds tested, IDA was the only substrate for the enzyme. Optimum activity of IDA-DH was found at pH 8.5 and 25°C, respectively, and the Km for IDA was found to be 8mM. Activity of the membrane-bound enzyme was inhibited by KCN, antimycine and dibromomethylisopropyl-benzoquinone. When inhibited by KCN IDA-DH was able to reduce the artificial electron acceptor iodonitrotetrazolium (INT). It was possible to extract IDA-DH from the membranes with 2% cholate, to reconstitute the enzyme into soybean phospholipid vesicles and to obtain IDA-DH activity (more than 50% recovery) using ubiquinone Q1 as the intermediate electron carrier and INT as the final electron acceptor. Growth experiments with different substrates revealed that in all NTA-degrading strains tested both NTA monooxygenase and IDA-DH were only expressed when the cells were grown on NTA or IDA. Furthermore, in Cb. heintzii ATCC 29600 growing exponentially on succinate and ammonia, addition of 0.4 g l-1 NTA led to the induction of the two enzymes within an hour and NTA was utilized simultaneously with succinate. The presence of IDA-DH was confirmed in ten different NTA-degrading strains belonging to three different genera.
Similar content being viewed by others
Abbreviations
- cA:
-
component A
- cB:
-
component B
- DBMIB:
-
dibromomethylisopropyl-benzoquinone
- HEPES:
-
hydroxyethylpiperazinethanesulfonic acid
- IDA:
-
iminodiacetate, HN(CH2COOH)2
- IDA-DH:
-
iminodiacetate dehydrogenase
- INT:
-
iodonitrotetrazolium chloride
- NTA:
-
nitrilotriacetate, N(CH2COOH)3
- NTA-MO:
-
nitrilotriacetate monooxygenase
- PMS:
-
phenazine methosulphate
- SDS-PAGE:
-
sodium dodecylsulfate polyacrylamide gel electrophoresis
- Suc-DH:
-
succinate dehydrogenase
References
Anonymous (1928) International critical tables. McGraw-Hill Book Company, New York
Auling G, Busse H-J, Egli T, El-Banna T & Stackebrandt E (1993) Chelatobacter, gen. nov. and Chelatococcus, gen. nov., two novel genera of the alpha subclass of the Proteobacteria to accomodate the Gram-negative, obligately aerobic, nitrilotriacetate (NTA)-utilizing bacteria Chelatobacter heintzii, sp. nov., and Chelatococcus asaccharovorans, sp. nov., System. Appl. Microbiol. (in press)
BamforthCW & LargePJ (1977) Solubilization, partial purification, and properties of N-methylglutamate dehydrogenase from Pseudomonas aminovorans. Biochem. J. 161: 357–370
BaterAJ & VenablesWA (1977) The characterisation of inducible dehydrogenases specific for the oxidation of D-alanine, allohydroxyproline, choline and sarcosine as peripheral membrane proteins in Pseudomonas aeruginosa. Biochim. Biophys. Acta 468: 209–226
BellyRT, LauffJJ & GoodhueCT (1975) Degradation of ethylenediaminetetraacetic acid by microbial populations from an aerated lagoon. Appl. Microbiol. 29: 787–794
BoultonCA, HaywoodGW & LargePJ (1980) N-methylglutamate dehydrogenase, a flavoprotein purified from a new pink trimethylamine-utilizing bacterium. J. Gen. Microbiol. 117: 293–304
CrippsRE & NobleAS (1973) The metabolism of nitrilotriacetate by a Pseudomonad. Biochem. J. 136: 1059–1068
CroftsAR, MeinhardtSW, JonesKR & SnozziM (1983) The role of the quinone pool in the cyclic electron transfer chain of Rhodopseudomons sphaeroides. Biochim. Biophys. Acta 723: 202–218
CroftsAR & WraightCA (1983) The electrochemical domain of photosynthesis. Biochim. Biophys. Acta 726: 149–185
EgliT, BallyM & UetzT (1990) Microbial degradation of chelating agents used in detergents with special reference to nitrilotriacetic acid (NTA). Biodegradation 1: 121–132
EgliT, WeilenmannH-U, El-BannaT & AulingG (1988) Gramnegative, aerobic, nitrilotriactate-utilizing bacteria from wastewater and soil. Syst. Appl. Microbiol. 10: 297–305
EpsteinSS (1972) Toxicological and environmental implications on the use of nitrilotriacetic acid as a detergent builder. Int. J. Environ. Stud. 2: 291–311
FirestoneMK & TiedjeJM (1978) Pathway of degradation of nitrilotriacetate by a Pseudomonas species. Appl. Environ. Microbiol. 35: 955–961
FochtDD & JosephHA (1971) Bacterial degradation of nitrilotriacetic acid. Can. J. Microbiol. 17: 1553–1556
HarlowE & LaneD (1988) Antibodies. A Laboratory Manual. Cold Spring Harbour Laboratory, New York
IngledewWJ & PooleRK (1984) The respiratory chains of Escherichia coli. Microbiol. Rev. 48: 222–271
Jenal-Wanner U (1991) Anaerobic degradation of nitrilotriacetate in a denitrifying bacterium: Purification and characterization of the nitrilotriacetate dehydrogenase/nitrate reductase enzyme complex. Doctoral thesis, ETH No. 9531, Swiss Federal Institute of Technology, Zürich, Switzerland
KakiK, YamaguchiH, IguchiY, TeshimaM, ShirakashiT & KuriyamaM (1986) Isolation and characteristics of nitrilotriacetate degrading-bacteria. J. Ferment. Technol. 64: 103–108
KasprzakAA, PapasEJ & SteenkampDJ (1983) Identity of the subunits and the stoichiometry of prosthetic groups in trimethylamine dehydrogenase and dimethylamine dehydrogenase. Biochem. J. 211: 535–541
KeeseyJ (1987) Biochemica Information. Boehringer Mannheim Biochemicals, Indianapolis
Kemmler J (1992) Biochemistry of nitrilotriacetate degradation in the facultatively denitrifying bacterium TE11. Doctoral thesis, No. 9983, Swiss Federal Institute of Technology, Zürich, Switzerland
KitaK, VibatCRT, MeinhardtS, GuestJR & GennisRB (1989) One-step purification from Escherichia coli of complex II (Succinate:ubiquinone oxidoreductase) associated with succinate-reducible cytochrome b556. J. Biol. Chem. 264: 2672–2677
KnechtR & YoungJY (1986) Liquid chromatographic determination of amino acids after gas-phase hydrolysis and derivatization with (dimethylamino)-azobenzenesulfonyl chloride. Anal. Chem. 58: 2375–2379
LaemmliUK (1970) Cleavage of structural proteins during the assembly of bacteriophage T4. Nature 227: 680–685
MatsushitaK, NonobeM, ShinagawaE, AdachiO & AmeyamaM (1987) Reconstitution of pyrroloquinoline quinone-dependant d-glucose oxidase respiratory chain of Escherichia coli with cytochrome \(\varpi \) oxidase. J. Bacteriol. 169: 205–209
McFetersGA, EgliT, WilbergE, AlderA, SchneiderRP, SnozziM & GigerW (1990) Activity and adaptation of nitrilotriacetate (NTA)-degrading bacteria: field and laboratory studies. Water Res. 24: 875–881
MeibergJBM & HarderW (1979) Dimethylamine dehydrogenase from Hyphomicrobium X: purification and some properties of a new enzyme that oxidizes secundary amines. J. Gen. Microbiol. 115: 49–58
OlsiewskiPJ, KaczorowskiGJ & WalshC (1980) Purification and properties of d-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur protein from Escherichia coli B. J. Biol. Chem. 255: 4487–4494
PennoyerJD, OhnishiT & TrumpowerBL (1988) Purification and properties of succinate-ubiquinone oxidoreductase complex from Paracoccus denitrificans. Biochim. Biophys. Acta 935: 195–207
PetersonGL (1979) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochemistry 83: 346–356
PickaverAH (1976) The production of N-nitrosoiminodiacetate from nitrilotriacetete and nitrate by microorganism growing in mixed culture. Soil Biol. Biochem. 8: 13–17
ReddyTLP & WeberMM (1986) Solubilization, purification and characterization of succinate dehydrogenase from membranes of Mycobacterium phlei. J. Bacteriol. 167: 1–6
SchneiderR, ZürcherF, EgliT & HamerG (1989) Ion chromatography method for iminodiacetic acid determination in biological matrices in the presence of nitrilotriacetic acid. J. Chromat. 462: 293–301
SchneiderR (1988) Determination of nitrilotriacetate in biological matrices using ion exclusion chromatography. Anal. Biochem. 173: 278–284
TiedjeJM (1980) Nitrilotriacetate: Hindsight and gunsight. In: MakiAM, DicksonKL & CairnsJ (Eds) Biotransformation and Fate of Chemicals in the Aquatic Environment (pp 114–119) ASM, Washington
TiedjeJM, MasonBB, WarrenCB & MalekEJ (1973) Metabolism of nitrilotriacetate by cells of Pseudomonas species. Appl. Microbiol. 25: 811–818
TowbinH, StaechelinT & GordonG (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and applications. Proc. Natl. Acad. Sci. USA 76: 4350–4353
TrijbelsF & VogelsGD (1966) Degradation of allantoin by Pseudomonas acidovorans. Biochim. Biophys. Acta 113: 292–301
TushurashaviliPR, GavrikovaEV, LendenevAN & VinogradovAD (1985) Studies on the succinate dehydrogenating system. Isolation and properties of the mitochondrial succinate-ubiquinone reductase. Biochim. Biophys. Acta. 809: 145–159
Uetz T (1992) Biochemistry of nitrilotriacetate degradation in obligately aerobic, Gram-negative bacteria. Doctoral thesis, No. 9722, Swiss Federal Institute of Technology, Zürich, Switzerland
UetzT, SchneiderR, SnozziM & EgliT (1992) Purification and characterization of a two component monooxygenase that hydroxylates nitrilotriacetate (NTA) from Chelatobacter heintzii ATCC 29600. J. Bacteriol. 174: 1179–1188
VaradhacharyA & MaloneyPC (1990) A rapid method for reconstitution of bacterial membrane proteins. Mol. Microbiol. 4: 1407–1411
WannerU, KemmlerJ, WeilenmannH-U, EgliT, El-BannaT & AulingG (1990) Isolation and growth of a bacterium able to degrade nitrilotriacetate (NTA) under denitrifying conditions. Biodegradation 1: 31–41
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Uetz, T., Egli, T. Characterization of an inducible, membrane-bound iminodiacetate dehydrogenase from Chelatobacter heintzii ATCC 29600. Biodegradation 3, 423–434 (1992). https://doi.org/10.1007/BF00240364
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00240364