Summary
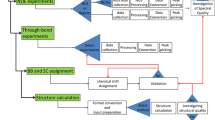
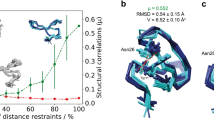
The AQUA and PROCHECK-NMR programs provide a means of validating the geometry and restraint violations of an ensemble of protein structures solved by solution NMR. The outputs include a detailed breakdown of the restraint violations, a number of plots in PostScript format and summary statistics. These various analyses indicate both the degree of agreement of the model structures with the experimental data, and the quality of their geometrical properties. They are intended to be of use both to support ongoing NMR structure determination and in the validation of the final results.
Similar content being viewed by others
References
Adobe Systems Inc (1985) PostScript Language Reference Manual, Addison-Wesley, Reading, MA, U.S.A.
AllenF.H. and JohnsonO. (1991) Acta Crystallogr., B47, 62–67.
BernsteinF.C., KoetzleT.F., WilliamsG.J.B., MeyerJr.E.F., BriceM.D., RodgersJ.R., KennardO., ShimanouchiT. and TasumiM. (1977) J. Mol. Biol., 112, 535–542.
BrüngerA.T. (1992) X-PLOR v. 3.1. A system for X-ray crystallography and NMR, Yale University Press, New Haven, CT, U.S.A.
ChazinW.J. (1992) Curr. Opin. Biotechnol., 3, 326–332.
CloreG.M., RobienM.A. and GronenbornA.M. (1993) J. Mol. Biol., 231, 82–102.
GonzalezC., RullmannJ.A.C., BonvinA.M.J.J., BoelensR. and KapteinR. (1991) J. Magn. Reson., 91, 659–664.
GüntertP., BraunW. and WüthrichK. (1991) J. Mol. Biol., 217, 517–530.
HavelT.F. and WüthrichK. (1984) Bull. Math. Biol., 46, 673–698.
HavelT.F. (1991) Prog. Biophys. Mol. Biol., 56, 43–78.
HochJ.C. (1991) In Computational Aspects of the Study of Biological Macromolecules by Nuclear Magnetic Resonance Spectroscopy (Ed.), HochJ.C., Plenum Press, New York, NY, U.S.A., pp. 253–267.
KabschW. and SanderC. (1983) Biopolymers, 22, 2577–2637.
KoradiR., BilleterM. and WüthrichK. (1996) J. Mol. Graph., 14, 51–55.
LaskowskiR.A., MacArthurM.W., MossD.S. and ThorntonJ.M. (1993) J. Appl. Crystallogr., 26, 283–291.
MacArthurM.W. and ThorntonJ.M. (1993) Proteins, 17, 232–251.
MacArthurM.W., LaskowskiR.A. and ThorntonJ.M. (1994) Curr. Opin. Struct. Biol., 4, 731–737.
MorrisA.L., MacArthurM.W., HutchinsonE.G. and ThorntonJ.M. (1992) Proteins, 12, 345–364.
RamachandranG.N., RamakrishnanC. and SasisekharanV. (1963) J. Mol. Biol., 7, 95–99.
SutcliffeM.J. (1993) In NMR of Macromolecules. A Practical Approach (Ed., RobertsG.C.K.), IRL Press, Oxford, U.K., pp. 359–392.
ThomasP.D., BasusV.J. and JamesT.L. (1991) Proc. Natl. Acad. Sci. USA, 88, 1237–1241.
VisH., MarianiM., VorgiasC.E., WilsonK.S., KapteinR. and BoelensR. (1995) J. Mol. Biol., 254, 692–703.
ZhaoD. and JardetzkyO. (1994) J. Mol. Biol., 239, 601–607.
Author information
Authors and Affiliations
Additional information
The AQUA and PROCHECK-NMR programs and operating instructions are available free of charge. The programs are supplied with script files for running on UNIX operating systems and can be obtained by anonymous ftp from 128.40.46.11 in directory pub/procheck. Further information can be obtained on http://www.biochem.ucl.ac.uk/~roman/procheck_nmr/procheck_nmr.html and http://www-nmr.chem.ruu.nl/users/rull/ aqua.html, or by sending e-mail to roman@bsm.bioc.ucl.ac.uk or rull@nmr.chem.ruu.nl.
R.A.L. and J.A.C.R. are joint first authors.
Rights and permissions
About this article
Cite this article
Laskowski, R.A., Rullmann, J.A.C., MacArthur, M.W. et al. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8, 477–486 (1996). https://doi.org/10.1007/BF00228148
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00228148