Abstract
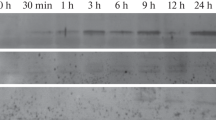
β-Conglycinin, the 7S storage protein of soybean, is expressed only in seeds and is regulated predominantly by gene transcription [5]. We applied an antisense strategy to modify expression of a β-glucuronidase (uidA or gusA) gene in seeds using a promoter from a β-conglycinin gene. Transgenic tobacco plants harboring the gusA gene under the control of the CaMV 35S promoter were retransformed with a gene construct comprising the β-conglycinin promoter fused to the gusA gene in the antisense orientation. Double transformants were regenerated and transformation was confirmed by Southern blot hybridization. Seed-specific repression of GUS activity was observed in lines containing high copy numbers of the antisense gusA transgene. Suppression of GUS activity was correlated with the amounts of (−) sense gusA transcript detected and concomitantly with a decrease in gusA transcript levels. Furthermore, the amount of suppression of GUS activity was greatest during mid to late stages of seed development, when expression of the α′ promoter is high. These results indicate that suppression of GUS activity is due to expression of the antisense gene.
Similar content being viewed by others
References
Bird CR, Ray JA, Fletcher JD, Boniwell JM, Bird AS, Teulieres C, Blain I, Bramley PM, Schuch W: Using antisense RNA to study gene function: inhibition of carotenoid biosynthesis in transgenic tomatoes. Bio/technology 9: 635–639 (1991).
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 (1976).
Cannon M, Platz J, O'Leary M, Sookdeo C, Cannon F: Organ-specific modulation of gene expression in transgenic plants using antisense RNA. Plant Mol Biol 15: 39–47 (1990).
Chen Z-L, Pan NS, Beachy RN: A DNA sequence element that confers seed-specific enhancement of a constitutive promoter. EMBO J 7: 297–302 (1988).
Chen Z-L, Schuler MA, Beachy RN: Functional analysis of regulatory elements in a plant embryo specific gene. Proc Natl Acad Sci USA 83: 8560–8564 (1986).
Coates JB, Medeiros JS, Thanh VH, Nielsen NC: Characterization of the subunits of β-conglycinin. Arch Biochem Biophys 243: 184–194 (1985).
Figurski DH, Helinski DR: Replication of an origincontaining derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76: 1648–1652 (1979).
Fraley RT, Rogers SG, Horsch RB, Eichholtz DA, Flick JS, Fink CL, Hoffman NL, Sanders PR: The SEV system: anew disarmed Ti plasmid vector system for plant transformation. Bio/technology 3: 629–635 (1985).
Hamilton AJ, Lycett GW, Grierson D: Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346: 284–287 (1990).
Hayford MB, Medford JI, Hoffman NL, Rogers SG, Klee HJ: Development of a plant transformation selection system based on expression of genes encoding gentamicin acetyltransferases. Plant Physiol 86: 1216–1222 (1988).
Horsch RB, Klee HJ: Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: Role of T-DNA borders in the transfer process. Proc Natl Acad Sci USA 83: 4428–4432 (1986).
Jefferson RA: Assaying chimeric genes in plants: the GUS fusion system. Plant Mol Biol Rep 5: 387–405 (1987).
Jefferson RA, Burgess SM, Hirsh D: β-Glucuronidase from Escherichia coli as a gene fusion marker. Proc Natl Soc Sci USA 83: 8447–8451 (1986).
Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions: β-glucuronidase as a sensitive and versatile fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B: Intracellular distribution of microinjected antisense oligonucleotides. Proc Natl Acad Sci USA 88: 2702–2706 (1991).
Matzke AJM, Stöger EM, Schernthaner JP, Matzke MA: Deletion analysis of a zein gene promoter in transgenic tobacco plants. Plant Mol Biol 14: 323–332 (1990).
Murashige T, Skoog F: A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 (1962).
Naito S, Dubé PH, Beachy RN: Differential expression of β-conglycinin α′ and β subunit genes in transgenic plants. Plant Mol Biol 11: 109–123 (1988).
Robert LS, Donaldson PA, Ladaique C, Altosaar I, Arnison PG, Fabijanski SF: Antisense RNA inhibition of β-glucuronidase gene expression in transgenic tobacco plants. Plant Mol Biol 13: 399–409 (1989).
Rodermel SR, Abbott MS, Bogorad L: Nuclear organelle interactions: Nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell 55: 673–681 (1988).
Rogers ST, Horsch RB, Fraley RT: Gene transfer in plants: Production of transformed plants using Ti plasmid vectors. Meth Enzymol 118: 627–640 (1986).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989).
Shortwell MA, Larkins BA: The biochemistry and molecular biology of seed storage proteins. In: The Biochemistry of Plants, vol. 15, pp. 297–345. Academic Press, New York (1989).
Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D: Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 334: 724–726 (1988).
Stockhaus J, Hofer M, Renger G, Westhoff P, Wydrzynski T, Willmitzer L: Anti-sense RNA efficiently inhibits formation of the 10 kd polypeptide of photosystem II in transgenic potato plants: analysis of the role of the 10 kd protein. EMBO J 9: 3013–3021 (1990).
Takayama KT, Inouye M: Antisense RNA. CRC Crit Rev Biochem Mol Biol 25: 155–184 (1990).
van derKrol AR, Lenting PE, Veenstra J, van derMeer IM, Koes RE, Gerates AGM, Mol JNM, Stuitje AR: An antisense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature 333: 866–869 (1988).
van derKrol AR, Mur LA, deLange P, Mol JNM, Stuitje AR: Inhibition of flower pigmentation by antisense CHS genes: promoter and minimal sequence requirements for the antisense effect. Plant Mol Biol 14: 457–466 (1990).
Visser PFG, Somhorst I, Kuipers GJ, Ruys NJ, Feenstra WJ, Jacobsen E: Inhibition of the expression of the gene for granule-bound starch synthase in potato by antisense constructs. Mol Gen Genet 225: 289–296 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fujiwara, T., Lessard, P.A. & Beachy, R.N. Seed-specific repression of GUS activity in tobacco plants by antisense RNA. Plant Mol Biol 20, 1059–1069 (1992). https://doi.org/10.1007/BF00028893
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00028893