Abstract
Immonium ions are commonly observed in the high energy fragmentation of peptide ions. In a MALDI-TOF/TOF mass spectrometer, singly charged peptides photofragmented with 157 nm VUV light yield a copious abundance of immonium ions, especially those from aromatic residues. However, their intensities may vary from one peptide to another. In this work, the effect of varying amino acid position, peptide length, and peptide composition on immonium ion yield is investigated. Internal immonium ions are found to have the strongest intensity, whereas immonium ions arising from C-terminal residues are the weakest. Peptide length and competition among residues also strongly influence the immonium ion production. Quantum calculations provide insights about immonium ion structures and the fragment ion conformations that promote or inhibit immonium ion formation.

ᅟ
Similar content being viewed by others
Introduction
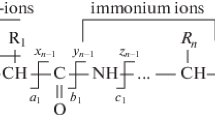
Immonium ions are small, single amino acid structures that result from peptide fragmentation in tandem mass spectrometry. The form of these ions is [NH2=CHR]+, where R is an amino acid side chain. Immonium ions associated with aromatic residues are particularly abundant [1]. High energy collision-induced dissociation (CID) was originally used to study these ions [1, 2], and various mechanisms of formation have been proposed [3–6]. For example, an N-terminal immonium ion can form from a rearrangement in the oxazolone ring of a b-type ion. An internal immonium ion can be created by first forming an a-type ion and subsequently releasing the N-terminal side of the ion [4]. Baldwin et al. found that factors such as location within the peptide influence the intensity of the immonium ion signal in mass spectra. They reported that the ion signals are strongest when the corresponding amino acid is located at the N-terminus and lowest at the C-terminus of peptides without an arginine or lysine [2]. Additionally, a competition between other residues within the peptide was discerned.
Since their observation can validate residues within a peptide, immonium ions are also relevant to peptide identification programs [7, 8]. For example, this might contribute to the analysis of antibodies. The complementarity determining regions (CDRs) of antibodies often contain hydrophobic residues, particularly those with aromatic side chains, which are involved in antigen binding [9]. Since aromatic residues produce abundant immonium ions, the observation of these small fragment ions can facilitate the recognition of CDR peptides.
In the present research, a matrix-assisted laser desorption ionization time-of-flight/time-of-flight (MALDI-TOF/TOF) mass spectrometer generates predominately singly charged ions. This instrument has been modified to achieve high energy fragmentation with a 157 nm vacuum ultraviolet (VUV) photodissociation laser [10]. Photodissociation provides an excellent opportunity to investigate trends associated with immonium ion formation because these ions are formed in abundance [11]. Ions associated with the aromatic residues histidine, tryptophan, tyrosine, and phenylalanine are the focus of this work. Variations in immonium ion yield due to position within a peptide, peptide length, and compositional effects are all examined. Additionally, theoretical calculations offer insights about the stability of various fragment structures and likely formation mechanisms.
Experimental
Materials
Peptides 1–8 in Table 1 were purchased from Peptide 2.0 (Chantilly, VA, USA). The residues of these peptides are commonly found in the complementarity determining regions of antibodies. Peptides 1–8 contain four aromatic residues in alternating positions. This enables studies of the positional dependence of immonium ion formation. Arginine is located at the N-terminus of four of the peptides and at the C-terminus of the other four. This enables the observation of both N- and C-terminal fragment ions. Peptides 9–40 were synthesized in-house. The matrix, α-cyano-4-hydroxycinnamic acid, was obtained from Fluka (St. Louis, MO, USA) and used for MALDI spotting with peptides. Rituximab was procured from Genentech (San Francisco, CA, USA). Angiotensin-I, bradykinin 2-9, trypsin from porcine pancreas, and S-methylisothiourea were supplied by Sigma (St. Louis, MO, USA).
Protein Digests
In addition to those directly synthesized, some peptides were obtained by digesting proteins with trypsin. Rituximab, a monoclonal antibody, was first reduced with 0.1 M dithiothreitol for 1 h at 56 °C. The reduced cysteines were alkylated with 0.1 M iodoacetamide in the dark for 30 min. The reaction was quenched with the addition of more dithiothreitol and spin filtered with ammonium bicarbonate solution to remove reagents. The antibody was then digested with trypsin (80% ammonium bicarbonate and 20% acetonitrile solution) in a Pressure BioSciences Inc. (South Easton, MA, USA) barocycler for 99 cycles at 37 °C in order to accelerate the digestion process. Each cycle was 50 s at 20 kpsi and 10 s at atmosphere. Guanidination labeling of lysines with S-methylisothiourea was performed to improve MALDI ionization [12]. A peptide library used in previous research contained peptides derived from tryptic digestion of ribosomal proteins [13, 14].
157 nm Photodissociation
For each synthesized peptide, 1 μL of approximately 500 μM peptide solution was deposited with 1 μL of 10 mg/mL of α-cyano-4-hydroxycinnamic acid in 50:50 water:acetonitrile with 0.1% trifluoroacetic acid. All antibody protein digests underwent reversed-phase liquid chromatography with an Eksigent Nano 2D LC coupled to an Eksigent Ekspot MALDI spotter (Framingham, MA, USA). This spotting procedure served to reduce the number of peptides per spot, minimizing ionization competition among peptides. A self-packed C18 capillary column was used to separate the mixture. The eluent was then mixed with matrix and spotted onto a MALDI plate.
As previously described, an ABI 4700 MALDI TOF-TOF mass spectrometer (Framingham, MA, USA) was modified to pass 157 nm VUV light from a Lambda Physik CompexPro laser (Santa Clara, CA, USA) through its collision cell [8]. The laser requires 5% fluorine in helium to produce the 157 nm light. As ions pass through the collision cell, they are irradiated with one light pulse. Resulting spectra include both PSD and photodissociation fragments. In Table 1, peptides 9–40 were analyzed on an ABI 4800 MALDI TOF-TOF mass spectrometer that was modified in a similar manner. Data from 1000 laser shots were averaged to create each spectrum.
Data Analysis
Data Explorer 4.6 software is provided with the ABI 4700 MALDI-TOF/TOF instrument. This software can calculate the percent abundance of each fragment relative to the height of the base peak. For the synthesized peptides, the relative percent abundance was averaged over two spectra, which improved data reproducibility. This analysis was also performed by calculating peak heights as a percentage of the total fragment ion intensity. Samples from the peptide library with low S/N were excluded from the data set.
Computer Simulations
Calculations were carried out using density functional theory (DFT) as implemented in the Jaguar 8.1 suite of ab initio quantum chemistry programs [15]. Geometry optimizations were performed with the hybrid functional B3LYP [16–19]. The 6-31G** basis set was used for all atoms [20, 21]. The energies of the optimized structures were reevaluated by additional single point calculations on each optimized geometry using Dunning’s correlation consistent triple-ζ basis set cc-pVTZ(-f) that includes a double set of polarization functions [22]. Vibration calculations were performed on all optimized structures to confirm that they were true minima that did not possess any imaginary frequencies. Vibrational/rotational/translational entropies of the molecule(s) were calculated using standard thermodynamic approximations. Changes in gas phase free energy ∆G were calculated as follows:
Where ∆G(gas) = change in gas phase free energy; ∆H(gas) = change in gas phase enthalpy; T = temperature (298.15 K); ∆S(gas) = change in gas phase entropy; ∆E(SCF) = self-consistent field energy, i.e., “raw” electronic energy as calculated at the triple-ζ level; ∆ZPE = change in vibrational zero point energy.
For reactions involving either a proton or hydrogen atom, we used Equation (3).
Here E(gas) is the gas-phase electronic energy, which is zero for a proton but non-zero for the hydrogen atom. The 5/2(RT) term is also included in estimations of reaction enthalpies. The translational entropy (S) for the hydrogen atom and proton was calculated as 26.04 eu using the Sackur-Tetrode equation.
To properly evaluate the energy of different structures, we attempted to locate the lowest energy conformer for each species, or at least one of the lowest energy conformers. The structures determined through our geometry optimizations only confirmed that we have obtained local minima, which will not necessarily be the global minima. We partially addressed this by using the Global-MMX program (GMMX) as employed by PCModel [23]. This conformational analysis employed the MMX force field [24] and generated a large number of reasonably low energy conformers. Although this analysis is useful for generating a small subset of reasonable structures, these structures and energies were not accurate enough for our analysis. These structures were therefore re-optimized with DFT. The gas phase free energy was calculated for a small set of the lowest energy DFT optimized structures (based on electronic energy at the double-ζ level), and the structure with the lowest gas phase free energy was then taken to be the global minimum. This procedure was employed for all structures except in instances where the number of structures to examine was particularly large. Conformational searches for open-shell species were performed on similar closed-shell species, and these cases are discussed in more detail in the Electronic Supplementary Material.
Results and Discussion
Two commonly investigated peptides, angiotensin-I and bradykinin 2-9, the sequences of which are DRVYIHPFHL and PPGFSPFR, were initially photodissociated in a MALDI-TOF/TOF mass spectrometer. Figure 1 shows expanded spectra of the low mass region for the two peptides. Since both sequences contain phenylalanine, these immonium ion intensities can be compared. Angiotensin-I produces a weak phenylalanine immonium ion with a 0.7% abundance relative to the base peak, whereas in bradykinin 2-9, this immonium ion is one of the most intense peaks in the spectrum with a relative abundance of 77%. This considerable difference in intensity demonstrates that position, sequence length, and/or peptide composition can impact the propensity to form these ions, and the following experiments consider these characteristics individually. After these factors are studied, angiotensin-I and bradykinin 2-9 will be further discussed.
Positional Dependence
To systematically investigate the effect of residue position on immonium ion yield, peptides 1–8 in Table 1 were MALDI ionized and photofragmented in the ABI 4700 TOF/TOF mass spectrometer. Figure 2 contains mass spectra of the four peptides (1–4) with an N-terminal arginine, and expanded views of the immonium ion regions of these spectra are displayed in Supplemental Figure S1. In Figure 2, large a-type ions terminated by histidine, phenylalanine, tryptophan, and tyrosine appear. For example, in addition to the intense a1 ions associated with arginine immonium ions, RHAFGWLY produces intense a2, a4, and a6 ions terminated by histidine, phenylalanine, and tryptophan, respectively. This is consistent with previous research [25–29]. Another observation is that the intensity of immonium ions varies across these peptides. The histidine immonium ion peak is substantially larger in the spectra of RYAHGFLW and RWAYGHLF than in that of RHAFGWLY and RFAWGYLH. The tryptophan immonium ion is much weaker in the RYAHGFLW spectrum than from the other three peptides. All four peptides yield relatively weak phenylalanine and tyrosine immonium ions, particularly when the residues are at the C-terminus. Mass spectra of peptides (5–8) with a C-terminal arginine are displayed in Figure 2, and their expanded low mass regions are in Supplemental Figure S2. The strong a-type ions that were present in the fragmentation of the N-terminal arginine peptides 1–4 are weaker or absent as expected. Instead, the C-terminal x-, y-, v-, and w-type fragments are formed. In these four peptides, the histidine and tryptophan immonium ions are more intense than corresponding tyrosine and phenylalanine ions, similar to the N-terminal arginine peptides.
Figure 3a summarizes the dependence of immonium ion signal on residue position for histidine, tryptophan, tyrosine, and phenylalanine in peptides with C-terminal arginines. Figure 3b contains analogous results for peptides with N-terminal arginines. In all cases, the blue curves denote the percent of the total fragment ion yield that is contributed by the histidine immonium ion. The red, green, and purple curves display analogous results for tryptophan, tyrosine, and phenylalanine, respectively. The abundance at each position reveals the probability of forming an immonium ion and how intense the ion is relative to the sum of all the peaks in a spectrum. A number of conclusions can be drawn. First, histidine forms the most intense immonium ion. Tryptophan is the second most abundant immonium ion followed by tyrosine and phenylalanine. Second, it is evident that certain positions within the peptide, such as at the C-terminus of N-terminal arginine peptides, produce weaker immonium ions than others. At this position, the histidine immonium ion exhibits a low relative abundance of only 1.2% whereas tryptophan, tyrosine, and phenylalanine are all below 0.2%. Third, histidine and tryptophan immonium ions both exhibit somewhat reduced intensities when located at or next to the N-terminus of the peptide. Tyrosine and phenylalanine are relatively weak in all positions. As an alternative approach to display the data, the immonium ion intensities were also plotted relative to the base peak values in Supplemental Figure S3. These graphs lead to similar conclusions.
Comparison of the immonium ion signals. (a) and (b) are peptides 1–8 in Table 1, which contain four aromatic residues; (c) and (d) are peptides 9–40 in Table 1 with one aromatic residue. The peptides in (a) and (c) contain a C-terminal arginine, whereas those in (b) and (d) contain an N-terminal arginine
A complication associated with the experiments just described is that the yield of immonium ions from a particular type of residue depends not only on the position of that residue in the peptide but also on what other residues are present in the sequence. As will be discussed in detail below, other aromatic residues within a peptide can compete for the available proton, thereby influencing the observed immonium ion intensities. In order to reduce this effect, peptides 9–40 were synthesized with only a single aromatic residue in various positions. The trends displayed in Figure 3c and d are similar to those in Figure 3a and b. The position of an aromatic residue strongly affects its immonium ion intensity.
Theoretical calculations provide insights into why histidine tends to form the most intense immonium ion. Figure 4 displays the lowest energy calculated structures for the immonium ions of phenylalanine, tyrosine, tryptophan, and histidine. In these structures, two different conformations are present: one in which the backbone atoms are in plane with the side chain ring and the other in which the backbone nitrogen is suspended over the side chain ring. Histidine exhibits this planar structure, whereas tryptophan, tyrosine, and phenylalanine adopt the nonplanar structure. Although nonplanar structures are visualized, alternative conformations where the backbone atoms are not positioned over the aromatic ring tend to be very close in energy (within ~2 kcal/mol), so this may not be a substantial characteristic. However, the key difference between histidine and the other amino acids is that histidine has a basic site in the side chain imidazole which, upon protonation, can engage in intramolecular H-bonding interactions, whereas phenylalanine, tyrosine, and tryptophan do not have an equivalent site. If histidine had the protonation site on the backbone imine and maintained the same H-bonding arrangement, the structure would be 1.33 kcal/mol higher in free energy. Although this energy difference is small, the important chemical interpretation is that the interaction of the side chain with the imine H, either genuine proton abstraction or strong H-bonding, is a critical stabilizing feature for this species. This is further shown by optimizing a structure where the immonium side chain has been rotated away from the imidazole side chain, which is 10.81 kcal/mol higher in free energy than the lowest energy H-bonded structure.
The energetics of immonium ion formation can also be partially justified by calculated proton affinities (PA) and gas-phase basicities (GB) as shown in Table 2. PA and GB are equivalent to –ΔH and –ΔG for protonation of a species. PAs and GBs were calculated for resultant immonium ions for all of the amino acids present in peptides 1–40. The trends for PA and GB are the same, so only the PAs will be discussed. The largest PA calculated is for arginine which, like histidine, preferentially undergoes protonation on its side chain as opposed to the imine nitrogen. The PAs then increase in the order Gly < Ala < Leu < Phe < Tyr < Trp < His < Arg. This order is the same as the order observed experimentally and computationally for the amino acids themselves [30, 31]. Interestingly, this trend also agrees with experimentally determined intensities of internal immonium ions, suggesting that PA is an important thermodynamic component for controlling which immonium ions are most likely to form. It has been shown that the PAs of amino acids are correlated with the intensity of molecular ions formed from MALDI experiments [32]. An exception to this trend in this work is arginine. Arginine ions have the greatest PA; much like histidine, they possess a basic side chain that can accept the proton and then engage in intramolecular H-bonding. While arginine a1 immonium ions form experimentally, they are not as intense as the other internal immonium ions. This is most likely the result of the arginine a1 ion being very susceptible to loss of NH3, and this a1-NH3 ion is indeed detected with high intensity (vide infra).
Peptides with an N-Terminal Arginine
The N-terminal arginine peptides have a clear pathway to internal immonium ions that has been previously outlined [4]. As noted above, large a-type ions terminated by each of the aromatic residues are abundantly formed. These can undergo a secondary reaction to break a peptide bond and form the immonium ion. Alternatively, it is well established that 157 nm photon absorption homolytically cleaves the Cα to CO bonds in a peptide [33]. Depending on the charge location, this results in a + 1 or x + 1 radical fragment ions, both of which can subsequently dissociate to more stable even-electron products, such as a-, x-, v-, and w-type ions. We now propose that a + 1 ions can also directly fragment to immonium ions as shown in Scheme 1.
Forming either an a-type ion or immonium ion from an a + 1 ion. The proton is not shown for the peptide ion, a + 1 ion or a-type ion. Just one a-type ion is displayed; others are shown in Scheme 2
As suggested in Scheme 1, an a + 1 ion may rearrange to produce an immonium ion or an even-electron a-type ion. Suppose that these are the only two possible products from an a + 1 precursor and that the probability of forming of an a + 1 ion terminated by a particular aromatic residue is independent of position. Based on these two assumptions, the sum of the intensities of the a-type ion and immonium ion formed from a specific aromatic residue should be the same at any position. Figure 5 shows the intensities of aromatic immonium ions formed at positions 2, 4, 6, and 8 and a-type ions resulting from cleavage C-terminal to these residues. The graphs on the left side of the figure were derived from peptides with four aromatic residues, whereas those on the right side originated from peptides with only a single aromatic residue. For positions 2, 4, and 6, it is apparent that the sum of the immonium and a-type ion intensities is approximately equal. This suggests that these ions are formed from a similar pathway such as that shown in Scheme 1. Note that for an eight-residue peptide a8 ions are not normally observed. As seen in Figure 5 when the only aromatic residue is present at position 4, 6, and 8, the immonium ion pathway is more favorable. On the other hand, for peptides with multiple aromatic residues, the an ion pathway is more favorable for all aromatic residues except for histidine. For example, if an a4 + 1 ion were formed from peptide 1 (RHAFGWLY), either the a4 ion or the phenylalanine immonium ion could be formed. However, the a4 + 1 ion includes arginine and histidine, the very high basicities of which would favor the production of a4 ions over immonium ions. This trend is consistent for each of the other a-type ions formed at tryptophan and tyrosine. Histidine, alternatively, has a higher basicity than the other aromatic residues and is strongly able to produce an immonium ion when located at positions 4, 6, and 8. For example, in peptide 2 (RYAHGFLW), the a4 + 1 ion proceeds to the histidine immonium ion rather than the a4 ion because of the higher basicity of the residue. In peptides with one aromatic residue, these effects are less pronounced because there is only one basic residue, arginine, and no other aromatic residue.
Immonium and a2 Ions
a2 Ions behave somewhat differently from a4 and a6 ions. As seen in Supplemental Figures S4 and S5, the a2 ion pathway is more favorable than any other a-type ion pathway and with nearly all aromatic residues, the a2 ion is more intense than the corresponding immonium ion. This increased a2 ion intensity suggests that its stability is enhanced, possibly due to a five-membered cyclic structure [34–36]. A strong a2-NH3 ion may result from an increased stability of the arginine side chain forming a seven-membered ring with the N-terminus [37].
There may be several reasons why an a2 ion is more intense than an aromatic immonium ion from the second position. The first is that the a2 ion could be more stable. DFT calculations were employed to investigate this possibility. The results of the calculations are briefly described here, while a more complete discussion can be found in Electronic Supplementary Material. Scheme 2 shows the possible reactions considered for a2 + 1 ions where the first residue was arginine, and the second was histidine, tryptophan, tyrosine, or phenylalanine (RH, RW, RY, and RF, respectively). To form an even electron a2 ion, a2 + 1 ions can expel a hydrogen atom by forming a double bond, either between the alpha and beta carbon, hereafter denoted as C=C, or between the alpha carbon and the amide nitrogen, hereafter denoted as N=C. In addition, a2 ions can cyclize to form an imidazolidinone structure, referred to as cyclic, as described previously [34–36]. The calculated thermodynamics are shown in Table 3. For all a2 ions considered, the N=C isomer was always significantly higher in energy than the C=C isomer. This can be readily explained by the fact that the C=C isomer is completely conjugated from the amide functionality to the aromatic side chain. The N=C isomer, on the other hand, has this conjugation interrupted by a methylene group. This is consistent with the work of Zhang et al [28, 29]. The cyclic a2 ions were essentially isoenergetic with the C=C structures in all examined cases and both are likely accessible. In all cases, however, forming the a2 ion was thermodynamically preferred over forming the immonium ion and a b1 radical. It is worth noting that of all the immonium ion formation pathways, it was most preferable to form histidine immonium ions, followed by tryptophan, and then tyrosine and phenylalanine. This again follows the experimentally observed order of ion intensities and agrees with the PA trend above.
A second potential reason why a2 + 1 ions may decay to a2 ions rather than immonium ions is that during 157 nm photodissociation, a peptide absorbs a 7.9 eV photon and fragments. If an a2 + 1 ion is formed, this ion could contain less internal excitation energy than a larger ion, preventing the immonium ion pathway. Formation of an a-type ion requires breaking a C–H or N–H bond [28], whereas formation of the immonium ion requires breaking the conjugated amide bond. Another possibility is that there is competition with the arginine at the N-terminus. A pathway described previously to an a1 ion indicates that proton affinity plays a critical role in determining what ion will be formed [6]. Since arginine has a higher proton affinity than the aromatic residues, an arginine a1 ion would be more favorable than the aromatic immonium ion. When aromatic residues are at other positions in these peptides, less intense a-type ions are observed, consistent with the more facile production of the immonium ion.
Peptides with a C-Terminal Arginine
C-terminal arginine peptides do not produce intense a-type ions, so creation of immonium ions requires an alternative mechanism. It has previously been argued that y-type ions can undergo a secondary reaction to break the Cα to CO bond and thereby produce immonium ions [4, 6]. Likewise in VUV photodissociation, x + 1 ions can rearrange to form x-, w-, and v-type ions [33]. As an alternative pathway, we propose that following the release of CO, instead of forming a v-type ion, the Cα to CO bond can be broken yielding an immonium ion as shown in Scheme 3.
Aromatic Residues at N- or C-Terminus
For peptides with a C-terminal arginine, immonium ions formed from aromatic residues at the N-terminus exhibit somewhat diminished intensities compared with positions 3 and 5. These a1 ions are not formed if the proton is sequestered on the C-terminal arginine. For histidine, this is less of a problem since the imidazole ring contains a basic protonation site that leads to the formation of strong a2 and b2 ions. As in the earlier discussion of aromatic residues next to N-terminal arginines, formation of these two ions potentially competes with formation of histidine immonium ions. For tryptophan, tyrosine, and phenylalanine, these residues lack a basic protonation site in their side chains, so the N-terminal amine will be the preferential protonation site [38]. If an a + 1 ion is the initial step to form this immonium ion, this fragment must contain the proton. Alternatively, immonium ions from internal residues initially form a + 1 or x + 1 radical ions. These radical ions are probably vibrationally excited with a mobilized proton that can form immonium ions more readily.
The reduced intensity for an immonium ion derived from an aromatic residue at the C-terminus likely involves several factors. Eight-residue peptides having N-terminal arginine do not form a8 ions. Consequently, C-terminal immonium ions would not form from a-type ion precursors. A mechanism for forming a phenylalanine immonium ion from a y1 ion has been proposed [4]. However, if arginine is at the N-terminus, relatively few y-type ions will be formed and particularly few y1 ions. Again, the only exception would be histidine, which is basic enough to form an intense y1 ion. DFT calculations on the lowest energy y1 ions show that histidine can be stabilized by an intramolecular H-bond between the imidazole and the ammonium ion; however, there are no other significant structural features in the other y1 ions. Because the protonation site of histidine is in its side chain imidazole, the interaction proposed by Harrison and coworkers to facilitate immonium ion formation will be unlikely, consistent with the weak immonium ion even for this residue [4]. Similarly, an x1 + 1 ion would likely be unable to form an immonium ion. This ion would have the same hydrogen bonds as the y1 ion that inhibits loss of the carboxylic acid to form the immonium ion.
To further support the observation of low intensity immonium ions from an amino acid at the C-terminus of a peptide, the arginine immonium ion at 129 Da was also investigated for peptides 1–8. However, this ion is typically weak with an average abundance of 5% relative to the base peak when arginine is at the N-terminus or less than 2% when at the C-terminus. Another arginine associated ion is at 112 Da, which is formed by the loss of ammonia from the immonium ion. The 112 Da ion is typically stronger than the immonium ion at 129 Da because a stable seven-membered ring is formed by losing ammonia [37]. When arginine is at the N-terminus, the average relative abundance of the 112 Da peak is 41%, and at the C-terminus, the abundance is 13%. With arginine at the C-terminus, y1 ions with considerable intensity are seen in Supplemental Figure S2. However, the loss of COOH needed to proceed to the immonium ion is likely inhibited by hydrogen bonds from the backbone carboxylic acid to the side chain guanidine group [39]. Additionally, this pathway is unlikely, which is consistent with histidine as noted above because both residues contain a side chain protonation site.
Length Dependence
Peptides of varying length were studied in order to learn how size impacts immonium ion yield. Previous research focused on small peptides because they tend to yield stronger immonium ions [2–5]. In this research, 238 peptides were generated from tryptic digestion of a variety of proteins, including hemoglobin, myoglobin, ubiquitin, cytochrome c, albumin, α-casein, carbonic anhydrase, ovalbumin, ribonuclease A, ribosomal proteins from Deinococcus radiodurans and E. coli, and the monoclonal antibody, Rituximab. Of the peptides identified, 90 contain histidine, 32 have tryptophan, 110 have tyrosine, and 121 have phenylalanine. The number of peptides with tryptophan is far less than the other aromatic residues because of its low natural abundance [40]. Figure 6 contains plots comparing the abundances of the four aromatic immonium ions relative to the base peak in each spectrum as a function of peptide length. In each graph, there is a clear decrease in immonium ion intensity as the peptide length increases. The large standard deviations calculated for each data set result from differences in sequence composition and in residue positions within the peptides. In an effort to reduce the scatter in the tryptophan plot, the immonium ion intensities were also displayed relative to the total fragment ion intensity as shown in Supplemental Figure S6; nevertheless, this produces a very similar trend. The conclusion from these data is that immonium ion signals decrease with peptide length. This is understandable from energetic considerations. The photon energy in the 157 nm laser is approximately 7.9 eV. Following photolytic bond cleavage, residual energy is available for vibrational excitation of the fragments. For smaller peptides, this residual energy will produce highly abundant low mass fragments. For larger peptides, on the other hand, there are more vibrational modes for the energy to be dispersed leading to less destructive fragmentation and fewer immonium ions.
Peptide Composition Dependence
As mentioned above, the presence of multiple aromatic residues in a single peptide can affect the immonium ion production. Figure 3a and b display data from peptides that each contained four aromatic residues, whereas Figure 3c and d show results for peptides with only a single aromatic residue. The latter two figures display a factor of two increase in the percent total fragment ion intensity for all immonium ions, as a result of reducing the competition with other residues. For example, the histidine immonium ion intensity from peptides with arginine at the C-terminus ranges from 4.7% to 6.7% in Figure 3a, but increases to 12.7%–18.2% in Figure 3c. In peptides with arginine at the N-terminus, the values range from 3.1%–6.3% in Figure 3b and increase to 6.1%–10.0% in Figure 3d. Similar results are obtained for the other aromatic residues. A second example of competition among residues for formation of immonium ions can be seen by comparing Figure 3c and d. The relative intensities of histidine, tryptophan, tyrosine, and phenylalanine immonium ions tend to be higher when arginine is at the C-terminus. As discussed in previous sections, when arginine is at the N-terminus, formation of a strong 112 Da ion competes with formation of aromatic immonium ions.
In order to further corroborate competition among residues, we investigated whether the peptides that produce the largest immonium ion signals are those with a single aromatic residue. In our 238 peptide library, 110 peptides contained tyrosine and 57 (51.8%) of these contained another aromatic residue. However, only three of the 21 peptides (14.3%) that produced an intense tyrosine immonium ion (above 50% of the base peak) contained another aromatic residue. Likewise, of the 121 peptides that contained phenylalanine, 60 (49.6%) contained another aromatic residue. However, of the 14 peptides that produced an intense phenylalanine immonium ion, only 1 (7.1%) contained another aromatic residue. This means that an intense tyrosine or phenylalanine immonium ion is less likely if another aromatic residue is in the sequence. The peptides in which tyrosine is present with another aromatic residue and still produces above 50% relative abundance are EFQTYFR, YAAHDENNEYK, and MESSAGTGFYYTTTK. For phenylalanine, the peptide is EFQTYFR. Although all four of these peptides contain another aromatic residue, they either contain two phenylalanine or tyrosine residues, or they have limited competition from the other residues within the sequence. Histidine and tryptophan were not included in this analysis because histidine typically has the most intense immonium ion regardless of peptide composition and there were not enough tryptophan containing peptides.
Having systematically investigated the factors that influence immonium ion intensity, we now reconsider data presented above for angiotensin-I and bradykinin 2-9. Based on their two sequences, DRVYIHPFHL and PPGFSPFR, the biggest contribution to the difference in phenylalanine immonium ion intensity is the sequence composition. Since the phenylalanines are located in the middle of the sequence, they do not suffer from the effects of being located at either termini or C-terminal to an arginine. The lengths of these peptides only differ by two residues, so no major contribution is expected from a length effect. The major difference is the sequence composition because angiotensin-I contains one phenylalanine, two histidines, and one tyrosine. With the presence of these other aromatic residues, the competition for producing a phenylalanine immonium ion is strongly impeded. On the other hand, bradykinin 2-9 contains only two phenylalanines and no other aromatic residues, so there is little competition with other amino acids.
Conclusions
A number of factors affect immonium ion intensity in 157 nm photodissociation experiments. Residue position, peptide length, and peptide composition were studied by synthesizing a variety of peptides and collecting peptides from tryptic digests of various proteins. It was found that residue position highly affects the immonium ion intensity. The positions of aromatic residues relative to other amino acids, such as arginine, are also an important factor that affects immonium ion intensity. Smaller peptides form more intense immonium ions than larger peptides. Finally, other residues within the peptide can reduce the intensity of an immonium ion because of competition for ionization. The results presented here will assist in identifying peptides that contain aromatic residues. An example would be peptides that contain the CDRs of antibodies.
Although early high-energy CID reports described abundant production of immonium ions [1–6], current proteomics experiments utilize low-energy CID, in which these ions are rarely observed both because they are not produced in abundance and because most experiments are performed with ion traps that have a low mass cut-off. In contrast, photodissociation in a MALDI-TOF/TOF mass spectrometer produces a plethora of detectable immonium ions. A future experiment that should be performed to further investigate these factors is to vary the intensity of the 157 nm laser. The abundance of each of the aromatic immonium ions might change as a result of increasing or decreasing the laser energy. Absorption of multiple photons can occur at higher laser energies that will increase the number of fragmentation events and complicate the trends observed in these experiments.
References
Falick, A.M., Hines, H.W., Medzihradszky, K.F., Baldwin, M.A., Gibson, B.W.: Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 4, 882–893 (1993)
Madden, T., Welham, J., Baldwin, M.A.: Factors affecting immonium ion intensities in the high-energy collision-induced decomposition spectra of peptides. Org. Mass Spectrom. 26, 443–446 (1991)
Dookeran, N., Yalcin, T., Harrison, A.G.: Fragmentation reactions of protonated α-amino acids. J. Mass Spectrom. 31, 500–508 (1996)
Ambihapathy, K., Yalcin, T., Leung, H.W., Harrison, A.G.: Pathways to immonium ions in the fragmentation of protonated peptides. J. Mass Spectrom. 32, 209–215 (1997)
Harrison, A.G., Csizmadia, I.G., Tang, T.H., Tu, Y.P.: Reaction competition in the fragmentation of protonated dipeptides. J. Mass Spectrom. 35, 683–688 (2000)
Paizs, B., Suhai, S.: Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 24, 508–548 (2005)
Wang, Y., Li, S.M., He, M.W.: Fragmentation characteristics and utility of immonium ions for peptide identification by MALDI-TOF/TOF-mass spectrometry. Chin. J. Anal. Chem. 42, 1010–1016 (2014)
Hohmann, L.J., Eng, J.K., Gemmill, A., Klimek, J., Vitek, O., Reid, G., Martin, D.B.: Quantification of the compositional information provided by immonium ions on a quadrupole-time-of-flight mass spectrometer. Anal. Chem. 80, 5596–5606 (2008)
Janeway Jr., C.A., Travers, P., Walport, M., Shlomchik, M.J.: Immunobiology: The Immune System in Health and Disease, 5th edn. Garland Science, New York (2001)
Zhang, L., Reilly, J.P.: Peptide photodissociation with 157 nm light in a commercial tandem time-of-flight mass spectrometer. Anal. Chem. 81, 7829–7838 (2009)
Liu, X., Li, Y.F., Bohrer, B.C., Arnold, R.J., Radivojac, P., Tang, H., Reilly, J.P.: Investigation of VUV photodissociation propensities using peptide libraries. Int. J. Mass Spectrom. 308, 142–154 (2011)
Beardsley, R.L., Reilly, J.P.: Optimization of guanidination procedures for MALDI mass mapping. Anal. Chem. 74, 1884–1890 (2002)
Zhang, L., Reilly, J.P.: Peptide de novo sequencing using 157 nm photodissociation in a tandem time-of-flight mass spectrometer. Anal. Chem. 82, 898–908 (2010)
Zhang, L., Reilly, J.P.: De Novo sequencing of tryptic peptides derived from Deinococcus radiodurans ribosomal proteins using 157 nm photodissociation MALDI TOF/TOF mass spectrometry. J. Proteome Res. 9, 3025–3034 (2010)
Jaguar, version 8.1, Schrodinger, LLC: New York, NY (2013)
Becke, A.D.: Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 38, 3098–3100 (1988)
Vosko, S.H., Wilk, L., Nusair, M.: Accurate spin-dependent electron liquid correlation energies for local spin density calculations: a critical analysis. Can. J. Phys. 58, 1200–1211 (1980)
Lee, C.T., Yang, W.T., Parr, R.G.: Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789 (1988)
Stephens, P.J., Devlin, F.J., Chabalowski, C.F., Frisch, M.J.: Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627 (1994)
Hehre, W.J., Ditchfield, R., Pople, J.A.: Self-consistent molecular orbital methods. XII. Further extensions of Gaussian-type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys. 56, 2257–2261 (1972)
Hariharan, P.C., Pople, J.A.: The influence of polarization functions on molecular orbital hydrogenation energies. Theor. Chim. Acta 28, 213–222 (1973)
Dunning Jr., T.H.: Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J. Chem. Phys. 90, 1007–1023 (1989)
PCMODEL, v.9.2, Serena Software: Box 3076, Bloomington, IN.
Gajewski, J.J., Gilbert, K.E., McKelvie, J.: In: Liotta, D. (ed.) Advances in Molecular Modeling, vol. 2, p. 65. JAI Press, Greenwich, CT (1990)
Chu, I.K., Rodriquez, C.F., Lau, T.C., Hopkinson, A.C., Siu, K.W.M.: Molecular radical cations of oligopeptides. J. Phys. Chem. B 104, 3393–3397 (2000)
Laskin, J., Yang, Z., Lam, C., Chu, I.K.: Charge-remote fragmentation of odd-electron peptide ions. Anal. Chem. 79, 6607–6614 (2007)
Ly, T., Julian, R.R.: Residue-specific radical-directed dissociation of whole proteins in the gas phase. J. Am. Chem. Soc. 130, 351–358 (2008)
Zhang, L., Cui, W., Thompson, M.S., Reilly, J.P.: Structures of α-type ions formed in the 157 nm photodissociation of singly-charged peptide ions. J. Am. Chem. Soc. 17, 1315–1321 (2006)
Zhang, L., Reilly, J.P.: Radical-driven dissociation of odd-electron peptide radical ions produced in 157 nm photodissociation. J. Am. Soc. Mass Spectrom. 20, 1378–1390 (2009)
Harrison, A.G.: The gas-phase basicities and proton affinities of amino acids and peptides. Mass Spectrom. Rev. 16, 201–217 (1997)
Uddin, K.M., Warburton, P.L., Poirier, R.A.: Comparisons of computational and experimental thermochemical properties of α-amino acids. J. Phys. Chem. B 116, 3220–3234 (2012)
Hatakeyama, M., Tachikawa, M.: Ab initio quantum chemical study on the mechanism of exceptional behavior of lysine for ion yields in MALDI—role of vibrational entropic contribution in thermally averaged proton affinities. J. Mass Spectrom. 46, 376–382 (2011)
Cui, W., Thompson, M.S., Reilly, J.P.: Pathways of peptide ion fragmentation induced by vacuum ultraviolet light. J. Am. Chem. Soc. 16, 1384–1398 (2005)
Aribi, H.E., Orlova, G., Rodriquez, C.F., Almeida, D.R.P., Hopkinson, A.C., Siu, K.W.M.: Fragmentation mechanisms of product ions from protonated tripeptides. J. Phys. Chem. B 108, 18743–18749 (2004)
Bythell, B.J., Maître, P., Paizs, B.: Cyclization and rearrangement reactions of an fragment ions of protonated peptides. J. Am. Chem. Soc. 132, 14766–14779 (2010)
Verkerk, U.H., Siu, C.-K., Steill, J.D., El Aribi, H., Zhao, J., Rodriquez, C.F., Oomens, J., Hopkinson, A.C., Siu, K.W.M.: a2 Ion derived from triglycine: an N1-protonated 4-imidazolidinone. J. Phys. Chem. Lett. 1, 868–872 (2010)
Bush, D.R., Wysocki, V.H., Scaraffia, P.Y.: Study of the fragmentation of arginine isobutyl ester applied to arginine quantification in Aedes aegypti mosquito excreta. J. Mass Spectrom. 47, 1364–1371 (2012)
Wu, Z., Fenselau, C.: Structural determinants of gas-phase basicities of peptides. Tetrahedron 49, 9197–9206 (1993)
Csonka, I.P., Paizs, B., Suhai, S.: Modeling of the gas-phase ion chemistry of protonated arginine. J. Mass Spectrom. 39, 1025–1035 (2004)
Neidhardt, F., Ingraham, J., Schaechter, M.: Physiology of the Bacterial Cell: A Molecular Approach. Sinauer Associates Inc., Massachusetts (1990)
Acknowledgment
The authors thank NIH and NSF for financial support under grants R01 GM103725 and CHE-1012855. M.H.B. thanks the Institute for Basic Science in Korea for support of this work (IBS-R10-D1).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 817 kb)
Rights and permissions
About this article
Cite this article
DeGraan-Weber, N., Ashley, D.C., Keijzer, K. et al. Factors Affecting the Production of Aromatic Immonium Ions in MALDI 157 nm Photodissociation Studies. J. Am. Soc. Mass Spectrom. 27, 834–846 (2016). https://doi.org/10.1007/s13361-015-1329-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1329-1