Abstract
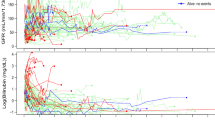
We are motivated by the chronic renal insufficiency cohort (CRIC) study to identify risk factors for renal progression in patients with chronic kidney diseases. The CRIC study collects two types of renal outcomes: glomerular filtration rate (GFR) estimated annually and end-stage renal disease (ESRD). A related outcome of interest is death which is a competing event for ESRD. A joint modeling approach is proposed to model a longitudinal outcome and two competing survival outcomes. We assume multivariate normality on the joint distribution of the longitudinal and survival outcomes. Specifically, a mixed effects model is fit on the longitudinal outcome and a linear model is fit on each survival outcome. The three models are linked together by having the random terms of the mixed effects model as covariates in the survival models. EM algorithm is used to estimate the model parameters, and the nonparametric bootstrap is used for variance estimation. A simulation study is designed to compare the proposed method with an approach that models the outcomes sequentially in two steps. We fit the proposed model to the CRIC data and show that the protein-to-creatinine ratio is strongly predictive of both estimated GFR and ESRD but not death.



Similar content being viewed by others
References
Anderson AH, Yang W, Hsu C-Y, Joffe MM, Leonard MB, Xie D, Chen J, Greene T, Jaar BG, Kao P et al (2012) Estimating gfr among participants in the chronic renal insufficiency cohort (cric) study. Am J Kidney Dis 60(2):250–261
Andrinopoulou E-R, Rizopoulos D, Takkenberg JJM, Lesaffre E (2014) Joint modeling of two longitudinal outcomes and competing risk data. Stat Med 33(18):3167–3178
Coresh J, Turin T, Matsushita K et al (2014) Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. JAMA 311(24):2518–2531
DeGruttola V, Tu XM (1994) Modelling progression of CD4-lymphocyte count and its relationship to survival time. Biometrics 50(4):1003–1014
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the em algorithm. J R Stat Soc Ser B (methodological) 39:1–38
Deslandes E, Chevret S (2010) Joint modeling of multivariate longitudinal data and the dropout process in a competing risk setting: application to ICU data. BMC Med Res Methodol 10(1):69
Diggle P, Kenward MG (1994) Informative drop-out in longitudinal data analysis. J R Stat Soc Ser C (Applied Statistics) 43(1):49–93
Efron B, Tibshirani RJ (1994) An introduction to the bootstrap. CRC Press, Boca Raton
Elashoff RM, Li G, Li N (2007) An approach to joint analysis of longitudinal measurements and competing risks failure time data. Stat Med 26(14):2813–2835
Feldman HI, Appel LJ, Chertow GM, Cifelli D, Cizman B, Daugirdas J, Fink JC, Franklin-Becker ED, Go AS, Hamm LL et al (2003) The chronic renal insufficiency cohort (cric) study: design and methods. J Am Soc Nephrol 14(suppl 2):S148–S153
Fine JP, Gray RJ (1999) A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 94(446):496–509
Guo X, Carlin BP (2004) Separate and joint modeling of longitudinal and event time data using standard computer packages. Am Stat 58(1):16–24
Hillis SL (1995) Residual plots for the censored data linear regression model. Stat Med 14(18):2023–2036
Hogan JW, Roy J, Korkontzelou C (2004) Handling drop-out in longitudinal studies. Stat Med 23(9):1455–1497
Hsieh F, Tseng Y-K, Wang J-L (2006) Joint modeling of survival and longitudinal data: likelihood approach revisited. Biometrics 62(4):1037–1043
Huang X, Li G, Elashoff RM, Pan J (2011) A general joint model for longitudinal measurements and competing risks survival data with heterogeneous random effects. Lifetime Data Anal 17:80–100
Klahr S, Levey AS, Beck GJ, Caggiula AW, Hunsicker L, Kusek JW, Striker G (1994) The effects of dietary protein restriction and blood-pressure control on the progression of chronic renal disease. N Engl J Med 330(13):877–884
Koller MT, Raatz H, Steyerberg EW, Wolbers M (2012) Competing risks and the clinical community: irrelevance or ignorance? Stat Med 31(11–12):1089–1097
Law M, Jackson D (2015) Residual plots for linear regression models with censored outcome data: a refined method for visualising residual uncertainty. Commun Stat Simul Comput. doi:10.1080/03610918.2015.1076470
Leung K-M, Elashoff RM, Afifi AA (1997) Censoring issues in survival analysis. Annu Rev Public Health 18(1):83–104
Li L, Hu B, Greene T (2009) A semiparametric joint model for longitudinal and survival data with application to hemodialysis study. Biometrics 65(3):737–745
Liu L, Ma JZ, O’Quigley J (2008) Joint analysis of multi-level repeated measures data and survival: an application to the end stage renal disease (esrd) data. Stat Med 27(27):5679–5691
Prentice RL, Kalbfleisch JD, Peterson AV Jr, Flournoy N, Farewell V, Breslow N (1978) The analysis of failure times in the presence of competing risks. Biometrics 34:541–554
Ratcliffe SJ, Guo W, Ten Have TR (2004) Joint modeling of longitudinal and survival data via a common frailty. Biometrics 60(4):892–899
Rizopoulos D (2010) Joint modelling of longitudinal and time-to-event data: challenges and future directions. In: 45th Scientific meeting of the Italian statistical society. Universitadi Padova, Padova
Rosansky SJ, Glassock RJ (2014) Is a decline in estimated gfr an appropriate surrogate end point for renoprotection trials & quest. Kidney Int 85(4):723–727
Schluchter MD (1992) Methods for the analysis of informatively censored longitudinal data. Stat Med 11(14–15):1861–1870
Song X, Davidian M, Tsiatis AA (2002) A semiparametric likelihood approach to joint modeling of longitudinal and time-to-event data. Biometrics 58(4):742–753
Stevens LA, Greene T, Levey AS (2006) Surrogate end points for clinical trials of kidney disease progression. Clin J Am Soc Nephrol 1(4):874–884
Taylor JMG, Park Y, Ankerst DP, Proust-Lima C, Williams S, Kestin L, Bae K, Pickles T, Sandler H (2013) Real-time individual predictions of prostate cancer recurrence using joint models
Trautmann H, Steuer D, Mersmann O, Bornkamp B (2014) truncnorm: truncated normal distribution. R package version 1.0-7
Tseng Y-K, Hsieh F, Wang J-L (2005) Joint modelling of accelerated failure time and longitudinal data. Biometrika 92(3):587–603
Tsiatis A (1975) A nonidentifiability aspect of the problem of competing risks. Proc Nat Acad Sci 72(1):20–22
Tsiatis AA, Davidian M (2001) A semiparametric estimator for the proportional hazards model with longitudinal covariates measured with error. Biometrika 88(2):447–458
Tsiatis AA, Davidian M (2004) Joint modeling of longitudinal and time-to-event data: an overview. Stat Sin 14(3):809–834
Tsiatis AA, DeGruttola V, Wulfsohn MS (1995) Modeling the relationship of survival to longitudinal data measured with error. Applications to survival and CD4 counts in patients with AIDS. J Am Stat Assoc 90(429):27
Vonesh EF, Greene T, Schluchter MD (2006) Shared parameter models for the joint analysis of longitudinal data and event times. Stat Med 25(1):143–163
Wang Y, Taylor JMG (2001) Jointly modeling longitudinal and event time data with application to acquired immunodeficiency syndrome. J Am Stat Assoc 96(455):895–905
Wilhelm S, Manjunath BG (2015) tmvtnorm: truncated multivariate normal and student t distribution. R package version 1.4-10
Williamson PR, Kolamunnage-Dona R, Philipson P, Marson AG (2008) Joint modelling of longitudinal and competing risks data. Stat Med 27(30):6426–6438
Wright JT Jr, Bakris G, Greene T, Agodoa LY, Appel LJ, Charleston J, Cheek D, Douglas-Baltimore JG, Gassman J, Glassock R et al (2002) Effect of blood pressure lowering and antihypertensive drug class on progression of hypertensive kidney disease: results from the aask trial. JAMA 288(19):2421–2431
Wu L, Liu W, Yi GY, Huang Y (2012) Analysis of longitudinal and survival data: joint modeling, inference methods, and issues. J Probab Stat 1–17:2012
Yu M, Law NJ, Taylor JM, Sandler HM (2004) Joint longitudinal-survival-cure models and their application to prostate cancer. Stat Sin 14(3):835–862
Acknowledgements
Wei Yang, Dawei Xie, Qiang Pan and Harold I. Feldman were supported by U01DK060990. Wensheng Guo was supported by R01GM104470. The authors thank the CRIC study patients, study coordinators, and investigators for their efforts. Other Grant Support: By the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (Cooperative Agreements U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK060980, U01DK060963, and U01DK060902) and in part by the following institutional Clinical Translational Science Awards from the National Center for Advancing Translational Sciences and other National Institutes of Health Grants: University of Pennsylvania (UL1TR000003, K01DK092353, and K24DK002651), Johns Hopkins University (UL1 TR-000424), University of Maryland General Clinical Research Center (M01RR-16500), Clinical and Translational Science Collaborative of Cleveland (UL1TR000439), Michigan Institute for Clinical and Health Research (UL1TR000433), University of Illinois at Chicago (UL1RR029879), Tulane University Translational Research in Hypertension and Renal Biology (P30GM103337), and Kaiser Permanente National Institutes of Health/National Center for Research Resources University of California, San Francisco Clinical and Translational Science Institute (UL1RR-024131).
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: Covariance Matrix for the Joint Distribution
According to models (1) and (2), we have
We suppress the subscript i for clarity.
Appendix 2: Expectation of the Sufficient Statistics
We first derive the expectations assuming both \(x_1\) and \(x_2\) are observed. We will then revise the formula accounting for the scenario when either or both \(x_1\) or \(x_2\) are censored. We use \(Y_{comp}\) to denote the full data including \(\mathbf {y}, x_1\) and \(x_2\).
According to models (1) and (2) and the covariance structure in the appendix, we have
where
and
\(E\left( {b{b^T}|{Y_{comp}},\hat{\theta }} \right) \) can then be calculated as
In the case when either or both \(x_1\) and \(x_2\) are censored,\(E(b | Y _{obs}, \hat{\theta })\) and \(E(b b^T | Y _{obs}, \hat{\theta })\) can be calculated as follows. We have
and
where \(\hat{b} = E\left( {b|Y_{comp},\hat{\theta }} \right) \). The last step follows because \(Var\left( {b|Y_{comp},\hat{\theta }} \right) \) is not a function of \(Y_{comp}\).
So the next step is to calculate \(E\left( {\hat{b}{{\hat{b}}^T}|Y_{obs},\hat{\theta }} \right) \). Since
we need to calculate \(E( {{x_1}|Y_{obs},\hat{\theta }})\), \(E( {{x_2}|Y_{obs},\hat{\theta }})\), \(E( {{x_1 ^2}|Y_{obs},\hat{\theta }})\), \(E( {{x_2 ^2}|Y_{obs},\hat{\theta }})\) and \(E( {{x_1 x_2}|Y_{obs},\hat{\theta }})\). See next section for the details of how to calculate these expectations under different scenarios. The calculation of \(E(\epsilon \epsilon ^T | Y_{obs}, \hat{\theta })\), \(E( {r_j^2|{Y_{obs}},\hat{\theta }} ),j=1,2\) can be done in a similar fashion.
To calculate \(E( x_jb|{Y_{obs}},\hat{\theta })\), \(j=1,2\), we have
Again see next section for the calculation of \(E(x_1^2|Y_{obs}, \hat{\theta })\), \(E(x_2^2|Y_{obs}, \hat{\theta })\) and \(E(x_1x_2|Y_{obs}, \hat{\theta })\).
To summarize, here are the formulas to calculate these quantities.
-
\(E(b_i|Y_{obs}, \hat{\theta }) = C_{12}C_{22}^{-1}E(O_i)\)
-
\(E(b_i b_i ^T|Y_{obs}, \hat{\theta }) = C_{12}C_{22}^{-1}E(O_iO_i^T)C_{22}^{-1}C_{12}^T+ \hat{D} - C_{12}C_{22}^{-1}C_{12}^T\)
-
\(E(\epsilon _i \epsilon _i ^T|Y_{obs}, \hat{\theta }) = F_{12}C_{22}^{-1}E(O_iO_i^T)C_{22}^{-1}F_{12}^T+ \hat{\sigma }^2 I - F_{12}C_{22}^{-1}F_{12}^T\)
-
\(E(r_{i,1}^2 |Y_{obs}, \hat{\theta }) = G_{12}C_{22}^{-1}E(O_iO_i^T)C_{22}^{-1}G_{12}^T+ \hat{s}_1^2 - G_{12}C_{22}^{-1}G_{12}^T\)
-
\(E(r_{i,2}^2 |Y_{obs}, \hat{\theta }) = H_{12}C_{22}^{-1}E(O_iO_i^T)C_{22}^{-1}H_{12}^T+ \hat{s}_2^2 - H_{12}C_{22}^{-1}G_{12}^T\)
-
\(E(x_{i,1}b_i|Y_{obs},\hat{\theta }) = C_{12}C_{22}^{-1}E(x_{i,1}O_i)\)
-
\(E(x_{i,2}b_i|Y_{obs},\hat{\theta }) = C_{12}C_{22}^{-1}E(x_{i,2}O_i)\)
where
-
\({C_{12}} = ( {\begin{array}{*{20}{c}} {\hat{D}{Z^T}}&{\hat{D}{{\hat{\lambda }}_1}}&{\hat{D}{{\hat{\lambda }}_2}} \end{array}} )\)
-
\({F_{12}} = ( {\begin{array}{*{20}{c}} {{{\hat{\sigma }}^2} I}&\mathbf {0}&\mathbf {0} \end{array}} )\)
-
\({G_{12}} = ( {\begin{array}{*{20}{c}} \mathbf {0}&{\hat{s}_1^2}&0 \end{array}} )\)
-
\({H_{12}} = ( {\begin{array}{*{20}{c}} \mathbf {0}&0&{\hat{s}_2^2} \end{array}} )\)
-
\({C_{22}} = \left( {\begin{array}{*{20}{c}} {Z\hat{D}{Z^T} + {{\hat{\sigma }}^2}I}&{}\quad {Z\hat{D}{{\hat{\lambda }}_1}}&{}\quad {Z\hat{D}{{\hat{\lambda }}_2}}\\ {\hat{\lambda }_1^T\hat{D}{Z^T}}&{}\quad {\hat{\lambda }_1^T\hat{D}{{\hat{\lambda }}_1} + \hat{s}_1^2}&{}\quad {\hat{\lambda }_1^T\hat{D}{{\hat{\lambda }}_2}}\\ {\hat{\lambda }_2^T\hat{D}{Z^T}}&{}\quad {\hat{\lambda }_2^T\hat{D}{{\hat{\lambda }}_1}}&{}\quad {\hat{\lambda }_2^T\hat{D}{{\hat{\lambda }}_2} + \hat{s}_2^2} \end{array}} \right) \)
-
\(O_i=\left( {\begin{array}{*{20}{c}} {y_i - T\hat{\alpha }}\\ {{x_{i,1}} - {W^T}{{\hat{\xi }}_1}}\\ {{x_{i,2}} - {W^T}{{\hat{\xi }}_2}} \end{array}} \right) \)
Note that \(E(\epsilon _i ^T \epsilon _i |Y_{obs}, \hat{\theta })\) can be calculated by summing over the diagonal elements of \(E(\epsilon _i \epsilon _i ^T|Y_{obs}, \hat{\theta })\), i.e., \(E(\epsilon _i ^T \epsilon _i |Y_{obs}, \hat{\theta })=trace\{E(\epsilon _i \epsilon _i ^T|Y_{obs}, \hat{\theta })\}\).
Appendix 3: Calculation of the First Two Moments of x Given \(Y_{obs}\)
There are three types of individuals in the observed data: 1) those whose \(x_1\) are observed but not \(x_2\), i.e., \(x_1 < c, x_2 > c\); 2) those whose \(x_2\) are observed but not \(x_1\), i.e., \(x_1 > c, x_2 < c\); and 3) neither \(x_1\) nor \(x_2\) is observed, i.e, \(x_1> c, x_2 > c\). We now derive \(E( {{x_1}|Y_{obs},\hat{\theta }})\), \(E( {{x_2}|Y_{obs},\hat{\theta }})\), \(E( {{x_1 ^2}|Y_{obs},\hat{\theta }})\), \(E( {{x_2 ^2}|Y_{obs},\hat{\theta }})\) and \(E( {{x_1 x_2}|Y_{obs},\hat{\theta }})\) for each of the three scenarios.
1.1 Scenario I: \(x_2\) Alone is Censored
For the first scenario in which \(x_1\) is observed and \(x_2\) is censored, we have \(Y_{obs}=(\mathbf {y},x_1,x_2>c)\) and
-
\(E( {{x_1}|Y_{obs},\hat{\theta }})=x_1\)
-
\(E( {{x_1 ^2}|Y_{obs},\hat{\theta }})=x_1^2\)
-
\(E( {{x_2}|Y_{obs},\hat{\theta }}) = E(x_2|\mathbf {y},x_1,x_2>c, \hat{\theta })\)
-
\(E( {{x_1 x_2}|Y_{obs},\hat{\theta }})=x_1E( {{x_2}|\mathbf {y},x_1,x_2>c,\hat{\theta }})\)
-
\(E( {{x_2 ^2}|Y_{obs},\hat{\theta }}) = E^2(x_2|\mathbf {y},x_1,x_2>c, \hat{\theta }) + Var(x_2|\mathbf {y},x_1,x_2>c, \hat{\theta })\)
Because
and
we have
and
where \(K_{12} = (\hat{\lambda }_2 ^T D \hat{\lambda }_1, \hat{\lambda }_2 ^T D Z^T )\) and \(K_{22} = \left( {\begin{array}{*{20}{c}} {\hat{\lambda }_1^T \hat{D}{\hat{\lambda }_1} + \hat{s}_1^2}&{}{\hat{\lambda }_1^T \hat{D}{Z^T}}\\ {Z \hat{D}{\hat{\lambda }_1}}&{}{Z \hat{D}{Z^T} + {\hat{\sigma }^2}I} \end{array}} \right) \).
So the calculation of \( E(x_2|\mathbf {y},x_1,x_2>c, \hat{\theta })\) and \( Var(x_2|\mathbf {y},x_1,x_2>c, \hat{\theta })\) can be done based on the truncated normal distribution property. In R, this can be done easily using the truncnorm package [31].
1.2 Scenario II: \(x_1\) Alone is Censored
Similarly, for the second scenario in which \(x_2\) is observed and \(x_1\) is censored, we have \(Y_{obs}=(\mathbf {y},x_1>c,x_2)\) and
-
\(E( {{x_2}|Y_{obs},\hat{\theta }})=x_2\)
-
\(E( {{x_2 ^2}|Y_{obs},\hat{\theta }})=x_2^2\)
-
\(E( {{x_1}|Y_{obs},\hat{\theta }}) = E(x_1|\mathbf {y},x_1>c,x_2, \hat{\theta })\)
-
\(E( {{x_1 x_2}|Y_{obs},\hat{\theta }})=x_2E( {{x_1}|\mathbf {y},x_1>c,x_2,\hat{\theta }})\)
-
\(E( {{x_1 ^2}|Y_{obs},\hat{\theta }}) = E^2(x_1|\mathbf {y},x_1>c,x_2, \hat{\theta }) + Var(x_1|\mathbf {y},x_1>c,x_2, \hat{\theta })\)
To calculate \( E(x_1|\mathbf {y},x_2,x_1>c, \hat{\theta })\) and \( Var(x_1|\mathbf {y},x_2,x_1>c, \hat{\theta })\), we have
and
where \(P_{12} = (\hat{\lambda }_1 ^T D \hat{\lambda }_2, \hat{\lambda }_1 ^T D Z^T )\) and \(P_{22} = \left( {\begin{array}{*{20}{c}} {\hat{\lambda }_2^T \hat{D}{\hat{\lambda }_2} + \hat{s}_2^2}&{}{\hat{\lambda }_2^T \hat{D}{Z^T}}\\ {Z \hat{D}{\hat{\lambda }_2}}&{}{Z \hat{D}{Z^T} + {\hat{\sigma }^2}I} \end{array}} \right) \).
1.3 Scenario III: Both \(x_1\) and \(x_2\) are Censored
When both \(x_1\) and \(x_2\) are censored, i.e, \(x_1>c\) and \(x_2>c\),
-
\(E( {{x_1}|Y_{obs},\hat{\theta }}) = E(x_1|\mathbf {y},x_1>c,x_2>c, \hat{\theta })\)
-
\(E( {{x_1 ^2}|Y_{obs},\hat{\theta }}) = E^2(x_1|\mathbf {y},x_1>c,x_2>c, \hat{\theta }) + Var(x_1|\mathbf {y},x_1>c,x_2>c, \hat{\theta })\)
-
\(E( {{x_2}|Y_{obs},\hat{\theta }}) = E(x_2|\mathbf {y},x_1>c,x_2>c, \hat{\theta })\)
-
\(E( {{x_2 ^2}|Y_{obs},\hat{\theta }}) = E^2(x_2|\mathbf {y},x_1>c,x_2>c, \hat{\theta }) + Var(x_2|\mathbf {y},x_1>c,x_2>c, \hat{\theta })\)
-
\(E( {{x_1 x_2}|Y_{obs},\hat{\theta }})=Cov( {{x_1x_2}|\mathbf {y},x_1>c,x_2>c,\hat{\theta }}) + E(x_1|\mathbf {y},x_1>c,x_2>c, \hat{\theta })*E(x_2|\mathbf {y},x_1>c,x_2>c, \hat{\theta })\)
We have
and
where \(Q_{12} = (\hat{\lambda }_1 ^T D Z^T, \hat{\lambda }_2 ^T D Z^T )^T\) and \(Q_{22} = Z \hat{D}{Z^T} + {\hat{\sigma }^2}I\). R package tmvtnorm [39] can then be used to calculate the mean and covariance of the truncated variables.
Rights and permissions
About this article
Cite this article
Yang, W., Xie, D., Pan, Q. et al. Joint Modeling of Repeated Measures and Competing Failure Events in a Study of Chronic Kidney Disease. Stat Biosci 9, 504–524 (2017). https://doi.org/10.1007/s12561-016-9186-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12561-016-9186-4