Abstract
Embryogenesis is a dynamic process with an intrinsic variability whose understanding requires the integration of molecular, genetic, and cellular dynamics. Biological circuits function over time at the level of single cells and require a precise analysis of the topology, temporality, and probability of events. Integrative developmental biology is currently looking for the appropriate strategies to capture the intrinsic properties of biological systems. The “–omic” approaches require disruption of the function of the biological circuit; they provide static information, with low temporal resolution and usually with population averaging that masks fast or variable features at the cellular scale and in a single individual. This data should be correlated with cell behavior as cells are the integrators of biological activity. Cellular dynamics are captured by the in vivo microscopy observation of live organisms. This can be used to reconstruct the 3D + time cell lineage tree to serve as the basis for modeling the organism's multiscale dynamics. We discuss here the progress that has been made in this direction, starting with the reconstruction over time of three-dimensional digital embryos from in toto time-lapse imaging. Digital specimens provide the means for a quantitative description of the development of model organisms that can be stored, shared, and compared. They open the way to in silico experimentation and to a more theoretical approach to biological processes. We show, with some unpublished results, how the proposed methodology can be applied to sea urchin species that have been model organisms in the field of classical embryology and modern developmental biology for over a century.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The phenomenological and theoretical reconstruction of multiscale dynamics in animal embryogenesis: a general perspective
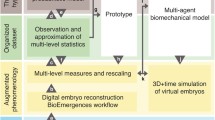
In an ideal scenario, morphogenetic events could be understood through a circular path integrating quantitative reconstructions, data analysis, and modeling (depicted in Fig. 1). The bottom-up digital reconstruction and analysis from the sub-cellular scale to the whole embryo level should be combined with a top-down approach. The latter includes the effect of high-level phenotypic features, such as tissue biomechanics on biochemical processes and on the regulation of gene expression. Events at the subcellular level modify the properties of cells and extracellular matrices, and the latter, in turn, modulates the biochemical activity of the cell [13, 74].
From in vivo and in toto observations to in silico experimentation, through the construction of digital embryos and the integration of quantitative data across spatio-temporal scales. From left to right. First column (light blue), volume rendering of experimental raw data displayed as snapshots taken from time-lapse sequences during Paracentrotus lividus embryo development. Second column (blue), surface rendering of digital embryos reconstructed from the raw data shown in the first column. Along the spatial scale, the microscopic level of the reconstructed data is at the single-cell scale (a single cell shown in yellow). The mesoscopic level corresponds to patterns/tissues (e.g., the Veg1 population in pink) and the macroscopic scale to the whole embryo (shown in purple). Third column (pink), quantitative description over time at the micro-, meso- and macroscopic levels extracted from digital embryos. Fourth column (green), theoretical modeling based on biological hypotheses and intended to predict the system behavior is compared with the quantitative observations. The cell is the integrator of subcellular activities including gene regulatory networks (GRNs), cell signaling and morphogen activity
Among all possible genetically and epigenetically encoded processes, cells divide, differentiate, and migrate in the context of their self-produced external environment [67, 102]. Cell features over time can be described with a set of parameters including the cell position, cell lineage, volume, surface, shape indexes, local convexity or concavity, nuclear/cytoplasmic ratio, cell neighborhood and surface of contact, cell polarization assessed by the asymmetric distribution of sub-cellular structures, intrinsic motility, and directionality. At the mesoscopic level, tissue patterning depends on cell displacements, cell–cell adhesion, and cell division characteristics. The emergence of the most macroscopic features shapes the whole embryo and provides global mechanical constraints. Changes in cell and tissue properties can be described by biomechanical models whose behavior depends on initial and boundary conditions given by the cellular environment [135]. A number of recent publications on different model organisms have focused on the mechanical properties of cells and their emergence from the interaction of cells with their environment [41, 46, 65, 66] and the interplay between mechanical constraints and gene expression patterning [13, 74, 114]. The biomechanical approach readily leads to the integration of the genetic and molecular basis of cell activities into the macroscopic deformation of tissues that shape the embryo [20].
Building multiscale models, integrating the biomechanics, and the molecular and genetic dynamics of embryonic development requires the quantitative description of cell behaviors. We review here how the state of the art reflects this paradigm, focusing on the reconstruction of digital specimens from in vivo and in toto imaging.
From in vivo imaging to modeling to understand animal embryogenesis: the premises
State of the art imaging techniques combined with the expression of fluorescent proteins provide time lapse recordings of the embryonic development of small transparent animal model organisms with resolution at the cellular level [69, 89]. The tridimensional images can be processed to reconstruct digital specimens composed of labeled volumes or surfaces corresponding to subcellular structures such as cell nuclei and membranes over time, and encompassing the clonal history of each cell. The digital reconstruction provides many advantages compared with raw 3D + time data. The visualization of biological structures (e.g., nuclei and membranes) in the digital specimen is simpler and can be combined with different color maps to highlight various features, select cells, and assess their clonal history (examples are shown for the sea urchin in Figs. 2 and 3). Even more importantly, digital embryos can be analyzed to quantify features at all scales.
Paracentrotus lividus embryo reconstruction at the 370-cell stage. Raw and reconstructed data displayed with the Mov-It interactive visualization software. Raw data with cell membranes (a) and nuclei (b) staining represented in volume rendering according to the color map displayed bottom right. Colored dots indicate the approximate nucleus center for each cell. Orthoslice in the x y plane for raw membranes (c) and nuclei (d), colored dots as in a,b; in the inset, orthoslices of raw data and their position in the volume indicated by blue lines. e Same as in c, with a cut of the segmented surfaces in addition. f Isosurfaces of segmented membranes. The different cell populations (i.e., small and large micromeres, Veg2 and Veg1 macromeres, and mesomeres) are displayed in different colors, color map described on the right. a–f Referential displayed bottom left. Scale bar 20 μm (Images from Duloquin, L., and Rizzi B., unpublished)
Cell clonal analysis in Paracentrotus lividus digital embryo. Segmentation of cell membranes represented as isosurfaces. a 32-cell stage, b 201-cell stage, c 334-cell stage, and d 545-cell stage. Scale bar 20 μm. e Flat representation of the cell lineage tree. Line segments between branching points represent the cell life time, branching points indicate mitoses. Color map (a-e) given by a set of colors associated with each cell at the 32-cell stage, color propagated along the cell lineage tree and thus revealing cell clones. This representation highlights the fact that very little cell dispersion occurs through the first 10 cell divisions (Reconstructions from live imaging from Rizzi B., unpublished)
A number of studies in different model organisms have more or less explicitly tackled the reconstruction of digital specimens from live observation. Pioneering work in the late 1970s [23, 128] led to the full reconstruction of the cell lineage tree of Caenorhabditis elegans [129]. This was obtained from the manual annotation of different embryos observed through Nomarski optics and was made possible thanks to the lineage invariance in Caenorhabditis elegans. Then, in the late 1990s, advances in microscopy and computation to handle high-dimensional data brought the first computer-aided cell tracking [124]. The authors used a new 3D multifocal time lapse system [55] to observe the early embryogenesis of Caenorhabditis elegans and the software Simi BioCell provided means for annotating and comparing the lineage trees of different embryos. The synergistic efforts of different disciplines brought further major advances. Developmental biology benefited from breakthroughs in fluorescent protein engineering [14, 16, 84, 85], microscopy imaging systems [34, 60, 107, 127, 130], image processing methods for cell segmentation and cell tracking [69, 90–92], the development of software for computer-aided data processing and visualization [26], and computer hardware for processing high dimensional datasets on computing grids [115].
From in vivo imaging to modeling to understand animal embryogenesis: reconstructing multiscale digital specimens
However, the reconstruction of animal embryogenesis from 3D + time imaging data remains a challenging approach, suffering from intrinsic limitations mainly related to the quality of the original data and the simultaneous requirement of automation and precision. The field of image processing has only recently turned to biological images, first dealing with 2D images, then 3D data, and then addressing one more level of complexity with 3D + time processing [93]. But even the most sophisticated algorithms are currently unable to distinguish objects or track them where the human eye cannot provide an accurate benchmark. So, the next breakthroughs in the field will have to originate from the biological side. In vivo 3D + time imaging has to deal with the trade-off between conflicting requirements including spatial and temporal resolution, signal-to-noise ratio, and photo damage. High spatial resolution and signal-to-noise ratio are needed to capture cell position and shape, but high temporal resolution is necessary to achieve single cell tracking. Further constraints are imposed by photo bleaching and photo damage, which limit the acquisition rate and overall imaging duration. In addition, fluorescent staining of subcellular structures in live model organisms is often non-homogeneous and adds artifacts in data acquisition, limiting the quality of the reconstruction, despite the development of sophisticated pre-processing methods [72, 134].
We can expect further improvements in fluorescent staining with more stable and brighter protein variants. Revealing contrasts intrinsic to the tissues such as the generation of harmonics by multiphoton illumination, has also proved to be useful [101]. When the cell density becomes too high, e.g., at late developmental stages, strategies to decrease the image complexity by providing color combinations and mosaic staining should be further developed. On the side of microscopy techniques, there is still room for improvement in depth imaging, spatial and temporal resolution, and signal-to-noise ratio. The recently developed single-plane illumination microscopy seems a valuable alternative to laser point-scanning microscopy, as it definitely improves the temporal resolution and the signal-to-noise ratio. But various imaging artifacts have not been solved yet and single-plane 2-photon excitation, which looks very promising, is still under development by physicists [62].
Except with ideal image data sets, e.g., highly contrasted objects at low density and moving slowly enough, state of the art image processing strategies are still far from providing error-free, fully automated processing of large 3D + time images. Every cell tracking error propagates to its progeny and impairs the reconstruction of cell clonal history. Similarly, shape segmentation inaccuracy adversely affects further analysis of the cells' characteristic features. Consequently, although protocols for 3D + time imaging of development based either on 2-photon or confocal laser scanning or on selective plane illumination microscopy have been described for several model organisms—Caenorhabditis elegans [44, 97], Ascidians [116–118], Drosophila melanogaster [105, 131], zebrafish [88], zebrafish and Drosophila [64], mouse [37, 100], and avian [73]—the most reliable results still come from manual reconstructions or extensively supervised and corrected data. In this context, the best results have been obtained for Nematoda and Ascidiacea embryos, thanks to their small cell number and largely invariant cell lineage that allows the matching of different specimens with precision at the single cell level (Table 1). Drosophila [35, 36, 68, 71, 80, 87, 109, 131] and zebrafish [63, 82, 101, 137] are more difficult to handle and cell tracking results obtained from state of the art 3D + time imaging data of developing embryos are suitable for average analysis but lack precision for cell clonal analysis. We review below the main results obtained for Nematodes which provides an example of error-free reconstruction of the cell lineage, and for Ascidians which gives an example of the digital representation of cell shapes combined with a quantitative description of cell interactions.
The digital reconstruction of Nematoda
The reconstruction of the cell lineage tree of Caenorhabditis elegans led to the development of dedicated software applications. The first available tools were designed to perform manual tracking and annotations. Worth mentioning, as it was pioneering in the field, is the development of 3D-DIASemb [50] for performing manual tracking, nuclear and membrane segmentation, and cell motion analysis from Nomarski video microscopy imaging of Caenorhabditis elegans up to the 28-cell stage. Simi BioCell, first mentioned in 1997, was used for manual tracking and annotation in Nematoda [58, 59], for combining the lineage tree with gene expression [96], and even lately for investigating cell lineages in the early mouse embryo [94]. Semiautomatic tracking with StarryNite [11] and AceTree [97] (available at http://waterston.gs.washington.edu/) provided the cell lineage tree of the Caenorhabditis elegans embryo up to 350 cells [8]. A digital atlas of the first larval stage (L1) with single-cell resolution was obtained from confocal images of 15 individual worms [78]. Gene expression and cell fate were then investigated [77] with the volume-object annotation system [108]. More recently, NucleiTracker4D (available at http://sourceforge.net/projects/nucleitracker4d/) was used for the semiautomatic tracking of nuclei imaged by laser scanning confocal microscopy of Caenorhabditis elegans development [39]. Further details concerning the cell clonal analysis in Caenorhabditis elegans can be found in [38, 44] and all the available data has been collected by the Nematode community on http://www.wormatlas.org/. It should be noted that the digital reconstruction performed so far on Nematoda has focused on displaying the gene regulatory network data according to cell position, but little integration has been achieved with cell behavior, and to our best knowledge, the assessment of cell shape through the segmentation of membrane images has not been addressed.
The digital reconstruction of Ascidiacea
As for the Nematodes, Ascidians exhibit a largely invariant cell lineage and digital embryos have been reconstructed from confocal images of fixed specimens at different stages of development. The most remarkable work has been collected in the Ascidian Network for In Situ Expression and Embryological Data (ANISEED) database [133] and the 3D Virtual Embryo software for visualization and interaction [132], both available through http://aniseed-ibdm.univ-mrs.fr/virtual_embryo.php. The 3D Virtual Embryo has been introduced as a tool for quantifying the geometry of cells and cell–cell contacts in an interactive three-dimensional environment and provides cell volumes, mathematical descriptors of cell shape, and cell–cell contacts. Initially used to describe the first developmental stages of Ciona intestinalis and a late 32-cell stage of Halocynthia roretzi, the concept has been extended to other Ascidian species. The ANISEED database integrating molecular, embryological, and anatomical data in the virtual embryo provides a new paradigm in the field of developmental biology. It should, however, be noted that so far, ANISEED has been based on the segmentation of fixed embryos. Further extending the concept will require the segmentation of live specimens. The Ascidian community has also contributed another interesting initiative. At first, dealing with raw data only, the Four-dimensional Ascidian Body Atlas, with interactive visualization of the development of Ciona intestinalis [57] is available online through http://chordate.bpni.bio.keio.ac.jp/faba/1.4/top.html. More recently, Hotta et al. achieved the manual segmentation of Ciona intestinalis tailbud embryos, and the interactive visualization of the data is available through 3D pdf files [98].
Reconstructing the multiscale dynamics of the sea urchin embryogenesis
The sea urchin as an animal model in experimental biology: historical perspective
The importance of the sea urchin as a model organism in biological investigations dates back to the nineteenth century and its characteristic features led to cornerstone discoveries in the history of embryology and cell biology. The transparency of the embryo allowed for the first time to observe the fusion between oocyte and sperm during the process of fertilization [52]. In 1892, blastomeres isolated at the 2-cell stage were shown to produce complete sea urchin larvae [24]. Such experimental possibilities made the sea urchin a very valuable model for investigating the components of individual variation. Pioneering studies on teratogenesis carried out by changing the chemical content of the sea urchin embryo helped to found the concept of embryonic induction [51]. The manipulation of double fertilized eggs [9] inspired the chromosome theory of inheritance and opened the way to the understanding of cancer as a genetic disease (English translation in [10]). Further work opened the way to modern Developmental Biology with the exploration of early development including gastrulation [56], cell mechanics underlying morphogenetic movements [18], and the mechanical properties of the sea urchin embryo at early stages of development [53, 54]. During the same period, Gustafson and Kinnander developed a technique allowing the gastrulation of the sea urchin Psammechinus miliaris to be analyzed for the first time, through time-lapse cinematography [42]. A few years later, the sea urchin was selected as a suitable animal model for large-scale analysis of gene expression regulation during early development [12]. More recently (2001), the discovery of cyclin and its role in cell cycle progression in the sea urchin [33] resulted in the award of the Nobel Prize in Physiology and Medicine to Tim Hunt (jointly with Leland H. Hartwell and Paul M. Nurse). Further information about the contribution of the sea urchin animal model to the foundation of paradigms in embryology and developmental biology can be found in [27].
The sea urchin in modern developmental biology
The sea urchin remains a leading model organism in developmental biology [4–6, 32], and the sea urchin community made a major breakthrough in building the architecture of the gene regulatory networks (GRN) underlying the embryo patterning and functional differentiation [86, 110]. The corresponding resources including protocols, database, and data annotation are publicly available (Table 2). However, cell lineage and biomechanics have only been scarcely addressed. The fate of embryonic cells was identified by means of fluorescent dyes [113, 120, 121] and uncaging methods [112], but very little work has been done to achieve in vivo and in toto 3D digital reconstructions from time-lapse imaging, and the resulting data have not yet permitted 3D + time quantitative analysis of cell behavior. Regarding the early development of Paracentrotus lividus, Luengo et al. [81] modeled cell geometries with a Voronoi diagram and compared results with membrane shapes obtained with a viscous watershed segmentation method [136]. Pastor et al. [106] designed a cell tracking method based on the introduction of a 4D tube-structuring element. Rubio et al. [119] merged lineage trees encompassing different periods of the sea urchin development using an error-tolerant graph matching technique. So far, however, these methods and the corresponding data have not led to new biological insights or any quantitative assessment of cell behavior. A quantitative 2D analysis of the mitotic gradient observed in different sea urchin species [1, 2, 103, 104] has been performed in the sand dollar Dendraster excentricus [25]. The quantitative assessment of cell behavior from time-lapse video microscopy imaging suggested a possible dependency of cell division timing and position on the cell volume between the fourth and the tenth cell cleavage. In this work, cell volumes were calculated indirectly from the measurement of cell diameter under the assumption of a spherical cell shape. Conclusions derived from 2D analysis might, however, be revisited at some point with more precise 3D data. This paradigm has been explored—although the corresponding data did not have single cell resolution—for the 3D reconstruction of adult sea urchin soft tissue from magnetic resonance imaging [138]. The method proposed for the inference of phylogenetic relationships was described together with a simple and attractive way to explore 3D structures interactively in digital specimens embedded in pdf files.
Reconstructing the digital sea urchin from 3D + time in toto imaging
The sea urchin has a number of desirable characteristics, making it a model of choice for in toto time-lapse microscopy imaging of the embryonic and larval stages. The embryo is easily accessible, relatively transparent and small, develops fast, grows to a fairly small number of cells and can be engineered to express fluorescent proteins. After overcoming the problem of the immobilization of the swimming blastula, the embryo can be imaged in toto for extended periods of time by two-photon laser scanning or selective plane illumination microscopy. The sea urchin is the ideal model for making a proof of concept for the reconstruction of the multiscale dynamics of a developing model organism from Deuterostomia, exploiting the unique data available in terms of the GRN architecture and dynamics.
We should, however, emphasize here that the task of long-term 3D + time imaging of the developing sea urchin and automated image processing to achieve cell tracking and shape segmentation is still subject to a number of limitations. These limitations are mainly on the biological side: highlighting structures without ectopic staining, e.g., staining membranes or nuclei without spurious cytoplasmic staining, immobilizing the specimen without mechanical deformation, compensating for light absorption and dispersion and thus loss of signal at depth, resolving individual nuclei throughout the entire volume including at late developmental stages, keeping a temporal resolution high enough to optimize the cell tracking outcome, e.g., ideally keeping the time step smaller than 2 min. Both 2-photon laser scanning and selective plane microscopy imaging are suitable, although they are based on quite different compromises, for the long-term 3D + time imaging of sea urchin embryos. It may soon be demonstrated that for specimens as small as sea urchin embryos, SPIM/DSLM offers the best conditions, provided that specimen mounting is improved compared with what is currently achieved [62].
Over the past few years, the BioEmergences platform (http://www.bioemergences.eu) and its interdisciplinary team have been exploring this paradigm, starting with the systematic reconstruction of the cell lineage tree of Paracentrotus lividus from in toto imaging and automated algorithmic processing. The reconstructed digital embryo consists of cell positions and shapes over time and cell clonal history. The corresponding data can be displayed and interactively explored, validated, and annotated with the visualization interface, Mov-IT, [101] (Figs. 2 and 3). The lineages of the different cell populations described in the early embryo according to [19] were highlighted from the 32-cell stage until blastula stages. These reconstructions provide precise measurements hitherto unavailable to model the dynamics at the microscopic scale and reconstruct the overall dynamics of the morphogenetic process at the macroscopic scale as well as possible. This is a most promising approach for the systematic identification of symmetry breaking or intra-individual and inter-individual variation. It would be a major paradigm change in the field of developmental biology, as it uses new theoretical frameworks to revisit classical embryology concepts.
When completed up to late developmental stages, the precise reconstruction of the cell's clonal history with associated shape, volume, and contact changes will contribute to our understanding of morphogenesis and differentiation processes [47–49] and answer a number of open questions. Digital specimens and the corresponding raw data should be made available to the entire community and scientists should all work to enrich a common database and contribute to the validation and correction of cell trajectories and cell shape. By sharing this 3D + time quantitative data for cohorts of individuals in defined genetic and environmental conditions, scientists will have the tools to bring a new perspective to classical questions of embryology.
First, a series of questions would be settled with the analysis of cell clonal history and cell fate such as for ectoderm/endoderm boundaries in the veg1 territory (reviewed by [32]) or the clonal origin of larval structures such as coelomic pouches. A second series of questions concerns the biomechanics of generic morphogenetic movements. The thickening of the vegetal plate during the ingression of the primary mesenchymal cells [28, 29] is described as a case of epithelial-mesenchymal transition, that should be compared to other similar processes [126]. The elongation and narrowing of the archenteron and the flattening of the epithelial cells in the wall gut rudiment [30], described in terms of the convergent extension movements [45], can be related to processes described in other model organisms [66, 67]. The Drosophila germ band extension [61], and the gastrulation and axis elongation in Ascidians [95], zebrafish [40, 70], avians [76, 122, 125], and mice [123] should, at some point, be quantified and compared. Achieving such a goal in the sea urchin will be insightful for a comparative study of morphogenetic processes and their evolutionary relationship.
Modeling morphogenetic processes in sea urchin development
The long-term goal of integrated modeling of the multiscale dynamics of sea urchin development will take advantage of the partial models established at different scales. The tentative modeling strategies available so far are scarce and much remains to be done in terms of comparing the models with quantitative data. A careful bibliographic search has brought to light four papers, all addressing early developmental steps and dealing respectively with: the biomechanical modeling of the primary invagination [21], the mechanical modeling of division asymmetry [3], the molecular modeling of cell proliferation [17] and the Boolean modeling of GRN dynamics [111].
Finite element modeling was used to discriminate between the possible mechanisms accounting for the primary invagination leading to the formation of the archenteron [21]. Hypothetical mechanisms driving invagination included at the time: apical constriction [7, 31], annular ring contraction [15, 31], cell tractoring [15], gel swelling [75], and apicobasal contraction [43]. But each of these mechanisms required different properties of the composite epithelial sheet. Further studies on Strongylocentrotus purpuratus embryos at the mesenchyme blastula stage [22] revealed that the extracellular matrix of the blastula wall was considerably stiffer than the cell layer. The authors then suggested that neither apical constriction nor annular ring contraction could drive invagination while all other hypotheses remained plausible.
Akiyama et al. developed an ad hoc mathematical model to reproduce the first four cleavages of the sea urchin egg, including the asymmetric divisions giving rise to micromeres [3]. The model established the planes of mitosis by assuming a chemotactic motion of centrosomes based on the diffusion of a chemical substance with repellent effects at the animal pole and attractive activity at the vegetal pole.
Ciliberto and Tyson [17] modified a previously developed mathematical model [99] based on molecular interactions underlying early embryonic cell-cycle control to investigate the hypothesis of a mitotic gradient along the animal-vegetal axis. Their results exhibited a good match with experimental data from [83] on Temnopleurus toreumaticus. They concluded that the mitotic wave was primarily attributable to differences in interdivision times in different parts of the embryo, although some subtle details of the wave were probably due to diffusive coupling between neighboring cells.
The transformation of the well-established GRN architecture into a predictive, dynamic Boolean computational model was a major recent breakthrough by the team led by Eric Davidson. This Boolean model computes spatial and temporal gene expression according to the GRN structure established for the embryonic development in the sea urchin. Direct comparison with experimental observations showed that the model predictively computed known gene expression patterns with remarkable spatial and temporal accuracy, thus validating the GRN architecture established so far.
All these models are intended to be both predictive and explanatory and to provide a formal basis to support or refute the intuition. They also tackle specific and limited scales and components of the system. Their integration in a multiscale modeling of morphogenesis will require a huge interdisciplinary and synergistic effort.
Conclusions
We have depicted an ideal scenario for a complex systems approach to morphogenetic processes underlying early animal development. The proposed strategy, based on the evidence that morphogenesis is a multiscale dynamic process, requires the integration of quantitative data across spatio-temporal scales into a yet-to-be-defined theoretical framework. A new interdisciplinary community coming from different perspectives and sharing the same goals is progressing fast. However, the overall conceptual and experimental landscape is still disparate, resembling a huge puzzle whose pieces do not yet fit together. Synergistic efforts by the scientific community are producing public databases of genome and GRN components for many model organisms. State-of-the-art microscopy imaging has made it possible to observe embryo development in small transparent organisms in toto, in vivo, with resolution at the cellular level. Software tools are being developed for visualizing, reconstructing and analyzing 3D + time image data. Computer hardware enables the processing of high-dimensional datasets on computing grids. Digital embryos can be directly manipulated and visualized in 3D to inspect and analyze cell clonal history and cell shape changes and cell–cell interactions over time. Much effort is being dedicated to the development of mathematical models of morphogenesis, based on either GRN architecture or the mechanical properties of tissue, for testing biological hypotheses and predicting the system's behavior. A new generation of digital resources anticipates the need to integrate multimodal and multiscale data and to provide tools for in silico experimentation. The availability of multimodal and multiscale quantitative data from the in vivo observation and reconstruction of cohorts of individuals should revolutionize practices in the field of developmental biology. Some major bottlenecks remain, related to the current limitations of long-term in vivo imaging and their consequences in terms of automated image processing. Imaging artifacts make 3D + time data from developing organisms difficult to process automatically. And the need for human supervision and time-consuming manual validation and correction is a major issue. The next breakthroughs may come from imaging techniques and may also rely on crowd sourcing for validating and correcting data. We also need new concepts in data analysis and theoretical modeling based on new representations of biological systems, faithful to biological observations but derived from formal descriptions to be shared by a new interdisciplinary community. In this context, the sea urchin is a model organism of choice for unprecedented achievements.
References
Agrell I (1964) Natural division synchrony and mitotic gradients in metazoan tissues. In: Zeuthen E (ed) Synchrony Cell Div. Growth. Interscience, New York, pp 32–67
Agrell I (1956) A mitotic gradient as the cause of the early differentiation in the sea urchin embryo. In: Wingstrand K (ed) Zoological Papers in Honour of B. Hanstrom, Lund, pp 27–34
Akiyama M, Tero A, Kobayashi R (2010) A mathematical model of cleavage. J Theor Biol 264:84–94. doi:10.1016/j.jtbi.2009.12.016
Angerer LM, Angerer RC (2003) Patterning the sea urchin embryo: gene regulatory networks, signaling pathways, and cellular interactions. Curr Top Dev Biol 53:159–198
Angerer LM, Angerer RC (2000) Animal-vegetal axis patterning mechanisms in the early sea urchin embryo. Dev Biol 218:1–12. doi:10.1006/dbio.1999.9553
Angerer LM, Oleksyn DW, Levine a M et al (2001) Sea urchin goosecoid function links fate specification along the animal-vegetal and oral-aboral embryonic axes. Development 128:4393–4404
Anstrom JA (1992) Microfilaments, cell shape changes, and the formation of primary mesenchyme in sea urchin embryos. J Exp Zool 264:312–322
Bao Z, Murray JI, Boyle T et al (2006) Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci U S A 103:2707–2712. doi:10.1073/pnas.0511111103
Boveri T (1904) Ergebnisse über die Konstitution der chromatischen Substanz des Zellkerns. Verlag von Gustav Fischer, Jena
Boveri T (2008) Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 121:1–84
Boyle TJ, Bao Z, Murray JI et al (2006) AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinforma 7:275. doi:10.1186/1471-2105-7-275
Britten RJ, Davidson EH (1969) Gene regulation for higher cells: a theory. Science 165:349–357
Brouzés E, Farge E (2004) Interplay of mechanical deformation and patterned gene expression in developing embryos. Curr Opin Genet Dev 14:367–374. doi:10.1016/j.gde.2004.06.005
Buckingham ME, Meilhac SM (2011) Tracing cells for tracking cell lineage and clonal behavior. Dev Cell 21:394–409. doi:10.1016/j.devcel.2011.07.019
Burke RD, Myers RL, Sexton TL, Jackson C (1991) Cell movements during the initial phase of gastrulation in the sea urchin embryo. Dev Biol 146:542–557
Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA (2010) Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev 90:1103–1163. doi:10.1152/physrev.00038.2009
Ciliberto A, Tyson JJ (2000) Mathematical model for early development of the sea urchin embryo. Bull Math Biol 62:37–59. doi:10.1006/bulm.1999.0129
Dan K, Okazaki K (1956) Cyto-embryological studies of sea urchins III. Role of the secondary mesenchime cells in the formation of the primitive gut in the sea urchin larvae. Biol Bull 110:29–42
Davidson EH (1989) Lineage-specific gene expression and the regulative capacities of the sea urchin embryo: a proposed mechanism. Development 105:421–445
Davidson LA, Von Dassow M, Zhou J (2009) Multi-scale mechanics from molecules to morphogenesis. Int J Biochem Cell Biol 41:2147–2162. doi:10.1016/j.biocel.2009.04.015
Davidson LA, Koehl MA, Keller R, Oster GF (1995) How do sea urchins invaginate? Using biomechanics to distinguish between mechanisms of primary invagination. Development 121:2005–2018
Davidson LA, Oster GF, Keller RE, Koehl MAR (1999) Measurements of mechanical properties of the blastula wall reveal which hypothesized mechanisms of primary invagination are physically plausible in the sea urchin Strongylocentrotus purpuratus. Dev Biol 209:221–238
Deppe U, Schierenberg E, Cole T et al (1978) Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A 75:376–380
Driesch H (1892) The potency of the first two cleavage cells in echinoderm development. Experimental production of partial and double formations. In: Willer BH, Openheimer JM (eds) Foundations of experimental embryology. Hafner, New York
Duncan REL, Whiteley AH (2011) The echinoid mitotic gradient: effect of cell size on the micromere cleavage cycle. Mol Reprod Dev 78:868–878. doi:10.1002/mrd.21373
Eliceiri KW, Berthold MR, Goldberg IG et al (2012) Biological imaging software tools. Nat Methods 9:697–710. doi:10.1038/nmeTh.2084
Ernst SG (2011) Offerings from an urchin. Dev Biol 358:285–294. doi:10.1016/j.ydbio.2011.06.021
Ettensohn CA (1985) Mechanisms of epithelial invagination. Q Rev Biol 60(3):289–307
Ettensohn CA (1984) Primary invagination of the vegetal plate during sea urchin gastrulation. Am Zool 24:571–588. doi:10.1093/icb/24.3.571
Ettensohn CA (1985) Gastrulation in the sea urchin embryo is accompanied by the rearrangement of invaginating epithelial cells. Dev Biol 112:383–390. doi:10.1016/0012-1606(85)90410-5
Ettensohn CA (1984) An analysis of invagination during sea urchin gastrulation. Yale University
Ettensohn CA, Sweet HC (2000) Patterning the early sea urchin embryo. Curr Top Dev Biol 50:1–44
Evans T, Rosenthal ET, Youngblom J et al (1983) Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33:389–396
Fischer RS, Wu Y, Kanchanawong P et al (2011) Microscopy in 3D: a biologist’s toolbox. Trends Cell Biol 21:682–691. doi:10.1016/j.tcb.2011.09.008.Microscopy
Fowlkes CC, Eckenrode KB, Bragdon MD et al (2011) A conserved developmental patterning network produces quantitatively different output in multiple species of Drosophila. PLoS Genet 7:e1002346. doi:10.1371/journal.pgen.1002346
Fowlkes CC, Hendriks CLL, Keränen SVE et al (2008) A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell 133:364–374. doi:10.1016/j.cell.2008.01.053
Garcia MD, Udan RS, Hadjantonakis AK, Dickinson ME (2011) Live Imaging of Mouse Embryos. Cold Spring Harb Protoc 2011:pdb.top104. doi: 10.1101/pdb.top104
Giurumescu CA, Chisholm AD (2011) Cell identification and cell lineage analysis. Meth Cell Biol 106:325–341. doi:10.1016/B978-0-12-544172-8.00012-8
Giurumescu CA, Kang S, Planchon TA et al (2012) Quantitative semi-automated analysis of morphogenesis with single-cell resolution in complex embryos. Development 139:4271–4279. doi:10.1242/dev.086256
Glickman NS (2003) Shaping the zebrafish notochord. Development 130:873–887. doi:10.1242/dev.00314
Gorfinkiel N, Schamberg S, Blanchard GB (2011) Integrative approaches to morphogenesis: lessons from dorsal closure. Genesis 49:522–533. doi:10.1002/dvg.20704
Gustafson T, Kinnander H (1956) Microaquaria for time-lapse cinematographic studies of morphogenesis in swimming larvae and observations on sea urchin gastrulation. Exp Cell Res 11:36–51. doi:10.1016/0014-4827(56)90188-4
Gustafson T, Wolpert L (1963) The cellular basis of morphogenesis and sea urchin development. Int Rev Cytol 15:139–214
Hardin J (2011) Imaging embryonic morphogenesis in C. elegans. MethodsCell Biol 106:377–412. doi:10.1016/B978-0-12-544172-8.00014-1
Hardin J (1989) Local shifts in position and polarized motility drive cell rearrangement during sea urchin gastrulation. Dev Biol 136:430–445. doi:10.1016/0012-1606(89)90268-6
Hardin J, Walston T (2004) Models of morphogenesis: the mechanisms and mechanics of cell rearrangement. Curr Opin Genet Dev 14:399–406. doi:10.1016/j.gde.2004.06.008
Hardin JD (1987) Archenteron elongation in the sea urchin embryo is a microtubule-independent process. Dev Biol 121:253–262
Hardin JD (1987b) The cellular mechanisms and mechanics of archenteron elongation in the sea urchin embryo. University of California
Hardin JD, Cheng LY (1986) The mechanisms and mechanics of archenteron elongation during sea urchin gastrulation. Dev Biol 115:490–501
Heid PJ, Voss E, Soll DR (2002) 3D-DIASemb: a computer-assisted system for reconstructing and motion analyzing in 4D every cell and nucleus in a developing embryo. Dev Biol 245:329–347. doi:10.1006/dbio.2002.0631
Herbst C (1893) Experimentelle Untersuchungen über den Einfluss der veränderten chemischen Zusammensetzung des umgebenden Mediums auf die Entwicklung der Thiere. II. Wierteres über die morphologische Wirkung der Lithiumsalze und ihre theoretische Bedeutung. Mitt D Zool Station Neapel 11:136–220
Hertwing O (1876) Beiträge zur Kenntniss des Bildung und Theilung des thierischen Eies. Morphologisches Jahrbücher 1:349–434
Hiramoto Y (1963) Mechanical properties of sea urchin eggs. I. Surface force and elastic modulus of the cell membrane. Exp Cell Res 32:59–75
Hiramoto Y (1963) Mechanical properties of sea urchin eggs. II. Changes in mechanical properties from fertilization to cleavage. Exp Cell Res 32:76–89
Hird SN, White JG (1993) Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol 121:1343–1355
Horstadius S (1953) Vegetalization of the sea-urchin egg by dinitrophenol and animalization by trypsin and ficin. J Embryol Exp Morphol 1:327–348
Hotta K, Mitsuhara K, Takahashi H et al (2007) A web-based interactive developmental table for the Ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn 236:1790–1805. doi:10.1002/dvdy.21188
Houthoofd W, Borgonie G (2007) The embryonic cell lineage of the nematode Halicephalobus gingivalis (Nematoda: Cephalobina: Panagrolaimoidea). Nematology 9:573–584
Houthoofd W, Jacobsen K, Mertens C et al (2003) Embryonic cell lineage of the marine nematode Pellioditis marina. Dev Biol 258:57–69. doi:10.1016/S0012-1606(03)00101-5
Huisken J, Swoger J, Del Bene F et al (2004) Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305:1007–1009. doi:10.1126/science.1100035
Irvine KD, Wieschaus E (1994) Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120:827–841
Keller P (2013) Imaging morphogenesis: technological advances and biological insights. Science 340(6137). doi:10.1126/science.1234168
Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK (2008) Reconstruction of Zebrafish early embryonic development by scanned light sheet microscopy. Science 322:1065–1069
Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK (2011) Digital scanned laser light-sheet fluorescence microscopy (DSLM) of zebrafish and Drosophila embryonic development. Cold Spring Harb Protoc 2011:1235–1243. doi:10.1101/pdb.prot065839
Keller R (2005) Cell migration during gastrulation. Curr Opin Cell Biol 17:533–541. doi:10.1016/j.ceb.2005.08.006
Keller R (2002) Shaping the vertebrate body plan by polarized embryonic cell movements. Science 298:1950–1954. doi:10.1126/science.1079478
Keller R, Davidson LA, Shook DR (2003) How we are shaped: the biomechanics of gastrulation. Differentiation 71:171–205. doi:10.1046/j.1432-0436.2003.710301.x
Keränen SVE, Fowlkes CC, Luengo Hendriks CL et al (2006) Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution II: dynamics. Genome Biol 7:R124. doi:10.1186/gb-2006-7-12-r124
Khairy K, Keller PJ (2011) Reconstructing embryonic development. Genesis 49:488–513. doi:10.1002/dvg.20698
Kimmel CB, Warga RM, Kane DA (1994) Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development 120:265–276
Knowles DW (2012) Three-dimensional morphology and gene expression mapping for the Drosophila blastoderm. Cold Spring Harb Protoc 2012:150–161. doi:10.1101/pdb.top067843
Krivá Z, Mikula K, Peyriéras N et al (2010) 3D early embryogenesis image filtering by nonlinear partial differential equations. Med Image Anal 14:510–526. doi:10.1016/j.media.2010.03.003
Kulesa PM, Bailey CM, Cooper C, Fraser SE (2010) In ovo live imaging of avian embryos. Cold Spring Harb Protoc 2010(6). doi:10.1101/pdb.prot5446
Kumar A, Shivashankar GV (2012) Mechanical force alters morphogenetic movements and segmental gene expression patterns during Drosophila embryogenesis. PLoS One 7:e33089. doi:10.1371/journal.pone.0033089
Lane MC, Koehl MAR, Wilt F, Keller R (1993) A role for regulated secretion of apical extracellular matrix during epithelial invagination in the sea urchin. Development 117:1049–1060
Lawson A, Schoenwolf GC (2001) Cell populations and morphogenetic movements underlying formation of the avian primitive streak and organizer. Genesis 29:188–195. doi:10.1002/gene.1023
Liu X, Long F, Peng H et al (2009) Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell 139:623–633. doi:10.1016/j.cell.2009.08.044
Long F, Peng H, Liu X et al (2009) A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat Methods 6:667–672. doi:10.1038/nmeth.1366
Longabaugh WJR, Davidson EH, Bolouri H (2005) Computational representation of developmental genetic regulatory networks. Dev Biol 283:1–16. doi:10.1016/j.ydbio.2005.04.023
Luengo Hendriks CL, Keränen SVE, Fowlkes CC et al (2006) Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol 7:R123. doi:10.1186/gb-2006-7-12-r123
Luengo-Oroz MA, Duloquin L, Castro C, et al. (2008) Can voronoi diagram model cell geometries in early sea-urchin embryogenesis? ISBI 2008. IEEE, pp 504–507
Luengo-Oroz MA, Rubio-guivernau JL, Faure E et al (2012) Methodology for reconstructing early zebrafish development from in vivo multiphoton microscopy. IEEE T Image Process 21:2335–2340
Masuda M, Sato H (1984) A synchronization of cell division is concurrently related with ciliogenesis in sea urchin blastulae. Dev Growth Differ 26:281–294
Mavrakis M, Pourquié O, Lecuit T (2010) Lighting up developmental mechanisms: how fluorescence imaging heralded a new era. Development 137:373–387. doi:10.1242/dev.031690
Mavrakis M, Rikhy R, Lilly M, Lippincott-Schwartz J (2008) Fluorescence imaging techniques for studying Drosophila embryo development. Curr Protoc Cell Biol 4(4.18). doi:10.1002/0471143030.cb0418s39
McClay DR (2011) Evolutionary crossroads in developmental biology: sea urchins. Development 138:2639–2648. doi:10.1242/dev.048967
McMahon A, Supatto W, Fraser S, Stathopoulos A (2008) Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science 322:1546–1550
Megason SG (2009) In toto imaging of embryogenesis with confocal time-lapse microscopy. In: Lieschke GJ, Oates AC, Kawakami K (eds) Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 317–332
Megason SG, Fraser SE (2003) Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech Dev 120:1407–1420. doi:10.1016/j.mod.2003.07.005
Meijering E, Dzyubachyk O, Smal I (2012) Methods for cell and particle tracking. Meth Enzymol 504:183–200
Meijering E, Dzyubachyk O, Smal I, Van Cappellen WA (2009) Tracking in cell and developmental biology. Semin Cell Dev Biol 20:894–902. doi:10.1016/j.semcdb.2009.07.004
Meijering E, Smal I, Dzyubachyk O, Olivo-Marin JC (2008) Time-lapse imaging. In: Wu Q, Merchant FA, Castleman KR (eds) Microscope image processing. Elsevier Academic Press, Waltham, pp 401–440
Mikut R, Dickmeis T, Driever W, et al. (2013) Automated Processing of Zebrafish Imaging Data: A Survey. Zebrafish. doi: 10.1089/zeb.2013.0886
Morris SA, Teo RTY, Li H et al (2010) Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci U S A 107:6364–6369. doi:10.1073/pnas.0915063107
Munro EM, Odell GM (2002) Polarized basolateral cell motility underlies invagination and convergent extension of the Ascidian notochord. Development 129:13–24
Murray JI, Bao Z, Boyle TJ et al (2008) Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods 5:703–709. doi:10.1038/NMETH.1228
Murray JI, Bao Z, Boyle TJ, Waterston RH (2006) The lineaging of fluorescently labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat Protoc 1:1468–1476. doi:10.1038/nprot.2006.222
Nakamura MJ, Terai J, Okubo R et al (2012) Three-dimensional anatomy of the Ciona intestinalis tailbud embryo at single-cell resolution. Dev Biol 372:274–284. doi:10.1016/j.ydbio.2012.09.007
Novak B, Tyson JJ (1993) Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J Cell Sci 106:1153–1168
Nowotschin S, Ferrer-Vaquer A, Hadjantonakis AK (2010) Imaging mouse development with confocal time-lapse microscopy. Methods Enzymol 476:351–377
Olivier N, Luengo-Oroz MA, Duloquin L et al (2010) Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329:967–971. doi:10.1126/science.1189428
Paluch E, Heisenberg C-P (2009) Biology and physics of cell shape changes in development. Curr Biol 19:R790–R799. doi:10.1016/j.cub.2009.07.029
Parisi E, Filosa S, Monroy A (1981) Spatial-temporal coordination of mitotic activity in developing sea urchin embryos. In: Haken H (ed) Chaos and Order in Nature. Springer, Berlin, pp 208–215
Parisi E, Filosa S, De Petrocellis B, Monroy A (1978) The pattern of cell division in the early development of the sea urchin, Paracentrotus lividus. Dev Biol 65:38–49
Parton RM, Vallés AM, Dobbie IM, Davis I (2010) Live cell imaging in Drosophila melanogaster. Cold Spring Harb Protoc 2010:pdb–top75. doi: 10.1101/pdb.top75
Pastor D, Luengo-Oroz MA, Lombardot B et al (2009) Cell tracking in fluorescence images of embryogenesis processes with morphological reconstruction by 4D-tubular structuring elements. EMBC 2009:970–973. doi:10.1109/IEMBS.2009.5333756
Pawley J (2006) Handbook of biological confocal microscopy. Springer, New York
Peng H, Long F, Myers EW (2009) VANO: a volume-object image annotation system. Bioinformatics 25:695–697. doi:10.1093/bioinformatics/btp046
Peng H, Long F, Zhou J et al (2007) Automatic image analysis for gene expression patterns of fly embryos. BMC Cell Biol 8(1):S7. doi:10.1186/1471-2121-8-S1-S7
Peter IS, Davidson EH (2011) A gene regulatory network controlling the embryonic specification of endoderm. Nature 474:635–639. doi:10.1038/nature10100
Peter IS, Faure E, Davidson EH (2012) Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci U S A 109:16434–16442. doi:10.1073/pnas.1207852109
Piston DW, Summers RG, Knobel SM, Morril JB (1998) Characterization of involution during sea urchin gastrulation using two-photon excited photorelease and confocal microscopy. Microsc Microanal 4:404–414
Ransick A, Davidson EH (1998) Late specification of Veg1 lineages to endodermal fate in the sea urchin embryo. Dev Biol 195:38–48. doi:10.1006/dbio.1997.8814
Rembold M, Loosli F, Adams RJ, Wittbrodt J (2006) Individual cell migration serves as the driving force for optic vesicle evagination. Science 313:1130–1134. doi:10.1126/science.1127144
Reuillon R, Chuffart F, Leclaire M, et al. (2010) Declarative task delegation in OpenMOLE. High Performance Computing and Simulation (HPCS), 2010 International Conference on. pp 55–62
Robin FB, Dauga D, Tassy O, et al. (2011a) Creating 3D digital replicas of Ascidian embryos from stacks of confocal images. Cold Spring Harb Protoc 2011:pdb–prot065862.
Robin FB, Dauga D, Tassy O, et al. (2011b) Time-lapse imaging of live Phallusia embryos for creating 3D digital replicas. Cold Spring Harb Protoc 2011:pdb–prot065847.
Robin FB, Dauga D, Tassy O, et al. (2011c) Imaging of fixed Ciona embryos for creating 3D digital replicas. Cold Spring Harb Protoc 2011:pdb–prot065854.
Rubio-Guivernau JL, Luengo-Oroz MA, Duloquin L et al (2009) Combining sea urchin embryo cell lineages by error-tolerant graph matching. EMBC 2009(2009):5918–5921. doi:10.1109/IEMBS.2009.5334851
Ruffins SW, Ettensohn CA (1993) A clonal analysis of secondary mesenchyme cell fates in the sea urchin embryo. Dev Biol 160:285–288. doi:10.1006/dbio.1993.1306
Ruffins SW, Ettensohn CA (1996) A fate map of the vegetal plate of the sea urchin (Lytechinus variegatus) mesenchyme blastula. Development 122:253–263
Sausedo RA, Schoenwolf GC (1993) Cell behaviors underlying notochord formation and extension in avian embryos: quantitative and immunocytochemical studies. Anat Rec 237:58–70. doi:10.1002/ar.1092370107
Sausedo RA, Schoenwolf GC (1994) Quantitative analyses of cell behaviors underlying notochord formation and extension in mouse embryos. Anat Rec 239:103–112. doi:10.1002/ar.1092390112
Schnabel R, Hutter H, Moerman D, Schnabel H (1997) Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev Biol 184:234–265. doi:10.1006/dbio.1997.8509
Schoenwolf GC, Alvarez IS (1989) Roles of neuroepithelial cell rearrangement and division in shaping of the avian neural plate. Development 106:427–439
Shook D, Keller R (2003) Mechanisms, mechanics and function of epithelial–mesenchymal transitions in early development. Mech Dev 120:1351–1383. doi:10.1016/j.mod.2003.06.005
So PTC, Dong CY, Masters BR, Berland KM (2000) Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng 2:399–429
Sulston JE, Horvitz HR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56:110–156
Sulston JE, Schierenberg E, White JG, Thomson JN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100:64–119
Sun C-K, Chu S-W, Chen S-Y et al (2004) Higher harmonic generation microscopy for developmental biology. J Struct Biol 147:19–30. doi:10.1016/j.jsb.2003.10.017
Supatto W, McMahon A, Fraser SE, Stathopoulos A (2009) Quantitative imaging of collective cell migration during Drosophila gastrulation: multiphoton microscopy and computational analysis. Nat Protoc 4:1397–1412. doi:10.1038/nprot.2009.130
Tassy O, Daian F, Hudson C et al (2006) A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol 16:345–358. doi:10.1016/j.cub.2005.12.044
Tassy O, Dauga D, Daian F et al (2010) The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res 20(10):1459–1568
Temerinac-Ott M, Ronneberger O, Ochs P et al (2012) Multiview deblurring for 3-D Images from light-sheet-based fluorescence microscopy. IEEE T Image Process 21:1863–1873
Trier SM, Davidson LA (2011) Quantitative microscopy and imaging tools for the mechanical analysis of morphogenesis. Curr Opin Genet Dev 21:664–670. doi:10.1016/j.gde.2011.08.005
Vachier C, Math CDM (2005) The viscous watershed transform. J Math Imaging Vis 22:251–267
Zanella C, Campana M, Rizzi B et al (2010) Cells segmentation from 3-D confocal images of early zebrafish embryogenesis. IEEE T Image Process 19:770–781. doi:10.1109/TIP.2009.2033629
Ziegler A, Faber C, Mueller S, Bartolomaeus T (2008) Systematic comparison and reconstruction of sea urchin (Echinoidea) internal anatomy: a novel approach using magnetic resonance imaging. BMC Biol 6:33. doi:10.1186/1741-7007-6-33
Acknowledgments
We thank Louise Duloquin for the time-lapse imaging of live sea urchin embryos and our collaborators at the BioEmergences platform for their contribution. This work was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche, the European Commission and the Region Ile de France. We thank Paul Bourgine for helpful discussions and Yannick Kergosien for critical reading of the manuscript. We apologize to the many authors whose work we were unable to cite owing to space constraints.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Rizzi, B., Peyrieras, N. Towards 3D in silico modeling of the sea urchin embryonic development. J Chem Biol 7, 17–28 (2014). https://doi.org/10.1007/s12154-013-0101-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12154-013-0101-x