Abstract
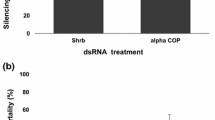
RNA interference (RNAi)-mediated gene silencing was explored for the control of sap-sucking pest Bemisia tabaci, commonly known as whitefly. dsRNAs and siRNAs were synthesized from five different genes – actin ortholog, ADP/ATP translocase, α-tubulin, ribosomal protein L9 (RPL9) and V-ATPase A subunit. A simplified insect bioassay method was developed for the delivery of ds/siRNA through the oral route, and efficacy was evaluated. ds/siRNA caused 29–97% mortality after 6 days of feeding. Each insect ingested nearly 150 nl of insect diet per day, which contained a maximum of 6 ng of RNA. Knocking down the expression of RPL9 and V-ATPase A caused higher mortality with LC50 11.21 and 3.08 μg/ml, respectively, as compared to other genes. Semi-quantitative PCR of the treated insects showed significant decrease in the level of RPL9 and V-ATPase A transcripts. siRNAs were found stable in the insect diet for at least 7 days at the room temperature. Phloem-specific expression of dsRNAs of RPL9 and V-ATPase A in transgenic plants for the protection against whiteflies might be an interesting application of this technology.




Similar content being viewed by others
Abbreviations
- ASAL:
-
Allium sativum agglutinin
- DMRT:
-
Duncan’s multiple range test
- EST:
-
expressed sequence tag; RNAi, RNA interference
- RPL9:
-
ribosomal protein L9
References
Baum JA, T. Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, et al. 2007 Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25 1322–1326
Blackburn MB, Domek JM, Gelman DB and Hu JS 2005 The broadly insecticidal Photorhabdus luminescens toxin complex a (Tca): Activity against the Colorado potato beetle, Leptinotarsa decemlineata, and sweet potato whitefly, Bemisia tabaci. J. Insect Sci. www.insectscience.org ISSN, 1536–2442
Brown JK and Czosnek H 2002 Whitefly transmission of plant viruses; in Advances in botanical research. plant virus vector interactions (ed) TR Plumb (Academic Press) vol. 36 pp 65–100
Bucher G, Scholten J and Klingler M 2002 Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 12 R85–R86
Buhtz A, Springer F, Chappell L, Baulcombe DC and Kehr J 2007 Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53 739–749
Byrne DN and Bellows Jr TS 1991 Whitefly biology. Annu. Rev. Entomol. 36 431–457
Carlini CR and Grossi-de-sa MF 2002 Plant toxic proteins with insecticidal properties. A review on their potentialities as bioinsecticides. Toxicon 40 1515–1539
Davidson EW, Patron RBR, Lacey LA, Frutos R, Vey A and Hedrix DL 1996 Activity of Natural toxins against the silverleaf whitefly, Bemisia argentifolii using a novel feeding bioassay system. Entomol. Exp. Appl. 79 25–32
Dutt U 2007 Mealy bug infestation in Punjab: Bt cotton falls flat. Environment News Service, 21 August (countercurrents.org)
Febvay G, Delobel B and Rahbe Y 1988 Influence of the amino acid balance on the improvement of an artificial diet for a biotype of Acyrthosiphon pisum (Homoptera: Aphididae). Can. J. Zool. 66 2449–2453
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC 1998 Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature (London) 391 806–811
Gatehouse AM, Powell KS, Peumans WJ, Van Damme EJM and Gatehouse JA 1995 Insecticidal properties of plant lectins: their potential in plant protection; in Lectins: Biomedical perspectives (eds) A Pusztai and S Bardocz (London: Taylor and Francis) pp 35–58
Ghanim M, Kontsedalov S and Czosnek H 2007 Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem. Mol. Biol. 37 732–738
Griebler M, Westerlund SA, Hoffmann KH and Meyering VM 2008 RNA interference with the allatoregulating neuropeptide genes from the fall armyworm Spodoptera frugiperda and its effects on the JH titer in the haemolymph. J. Insect Physiol. 54 997–1007
Huvenne H and Smagghe G 2010 Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 56 227–235
Jancovich JK, Davidson EW, Lavine M and Hendrix DL 1997 Feeding chamber and diet for culture of nymphal silverleaf whitefly, Bemisia argentifolii. J. Econ. Entomol. 90 628–633
Kehr J and Buhtz A 2008 Long distance transport and movement of RNA through the phloem. J. Exp. Bot. (Special Issue) 59 85–92
Kragler F 2010 RNA in the phloem: A crisis or a return on investment? Plant Sci. 178 99–104
Kumar M, Gupta GP and Rajam MV 2009 Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J. Insect Physiol. 55 273–278
Leshkowitz D, Gazit S, Reuveni E, Ghanim M, Czosnek H, McKenzie C, Robert Jr LS and Brown KJ 2006 0Whitefly (Bemisia tabaci) genome project: analysis of sequenced clones from egg, instar, and adult (viruliferous and non-viruliferous) cDNA libraries. BMC Genomics 7 79
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP and Chen XY 2007 Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25 1307–1313
Meyering-Vos M and Müller A 2007 RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J. Insect Physiol. 53 840–848
Mutti NS, Park Y, Reese JC and Reeck GR 2006 RNAi knockout of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J. Insect Sci. 6 38
Newmark PA, Reddien PW, Cebria F and Alvarado AS 2003 Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci USA 100 11861–11865
Pant BD, Buhtz A, Kehr J and Scheible WRD 2008 MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53 731–738
Price DRG and Gatehouse JA 2008 RNAi-mediated crop protection against insects. Trends Biotechnol. 26 393–400
Rajagopal R, Sivakumar S, Agrawal N, Malhotra P and Bhatnagar RK 2002 Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol. Chem. 277 46849–46851
Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B and Zhang W 2009 Developmental control of a Lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS ONE 4 e6225
Tomoyasu Y and Denell RE 2004 Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 214 575–578
Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP and Newcomb RD 2006 RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol. 15 383–391
Van Damme EJM, Peumans WJ, Barre A and Rouge P 1998 Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with divers biological role; Crit. Rev. Plant Sci. 17 575–692
Vermehren A, Qazi S and Trimmer BA 2001 The nicotinic alpha subunit MARA1 is necessary for cholinergic evoked calcium transients in Manduca neurons. Neurosci. Lett. 313 113–116
Virla EG, Casuso M and Frias EA 2010 A preliminary study on the effects of a transgenic corn event on the non target pest Dalbulus Maiid (Hemitera: Cicadeliidae). Crop Prot. 29 635–638
Acknowledgements
The authors are grateful to Council of Scientific and Industrial Research (CSIR), New Delhi, for the financial support. RT is grateful to Department of Science and Technology, New Delhi, for the JC Bose Fellowship. SKU is thankful to CSIR for the senior research fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Upadhyay SK Chandrashekar K, Thakur N, Verma PC, Borgio JF, Singh PK and Tuli R 2011 RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 36 153–161] DOI 10.1007/s12038-011-9009-1
MS received 29 July 2010; accepted 16 December 2010, ePublication: 14 March 2011 and Corresponding editor: Man Mohan Johri
Rights and permissions
About this article
Cite this article
Upadhyay, S.K., Chandrashekar, K., Thakur, N. et al. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci 36, 153–161 (2011). https://doi.org/10.1007/s12038-011-9009-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-011-9009-1