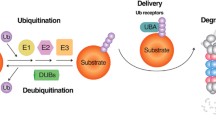
Abstract
Impairment in the clearance of misfolded proteins by functional proteins leads to various late-onset neurodegenerative diseases. Cell applies a strict quality control mechanism against malfunctioned proteins which may generate cellular proteoxicity. Under proteotoxic insults, cells immediately adopt two major approaches to either refold the misfolded proteinaceous species or degrade the unmanageable candidates. However, the main cellular proteostasis quality control mechanism is not clear. It is therefore important to understand the events and cellular pathways, which are implicated in the clearance of recalcitrant proteins. Ubiquitin proteasome system manages intracellular protein degradation. In this process, E3 ubiquitin ligase enzyme provides specificity for recognition of client proteins. In this review, we summarize various molecular approaches governed by E3 ubiquitin ligases in the degradation of aberrant proteins. A clear understanding of E3 ubiquitin ligases can offer a well tractable therapeutic approach against neurodegenerative diseases.



Similar content being viewed by others
References
Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416(6880):507–511. doi:10.1038/416507a416507a
Lorenzo A, Yankner B (1994) Beta-amyloid neurotoxicity requires fibril formation and is inhibited by congo red. Proc Natl Acad Sci U S A 91(25):12243–12250
Thomas T, Thomas G, McLendon C, Sutton T, Mullan M (1996) beta-Amyloid-mediated vasoactivity and vascular endothelial damage. Nature 380(6570):168–239. doi:10.1038/380168a0
Lindner AB, Demarez A (2009) Protein aggregation as a paradigm of aging. Biochim Biophys Acta 1790(10):980–996. doi:10.1016/j.bbagen.2009.06.005
Douglas P, Cyr D (2010) Interplay between protein homeostasis networks in protein aggregation and proteotoxicity. Biopolymers 93(3):229–265. doi:10.1002/bip.21304
Glickman M, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological Reviews 82(2):373–801. doi:10.1152/physrev.00027.2001
Glabe C (2006) Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiology of Aging 27(4):570–575. doi:10.1016/j.neurobiolaging.2005.04.017
Taylor JP, Hardy J, Fischbeck KH (2002) Toxic proteins in neurodegenerative disease. Science 296(5575):1991–1995. doi:10.1126/science.1067122296/5575/1991
Hartl F, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science (New York, NY) 295(5561):1852–1860. doi:10.1126/science.1068408
Morimoto R (1993) Cells in stress: transcriptional activation of heat shock genes. Science (New York, NY) 259(5100):1409–1419
Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27:437–496. doi:10.1146/annurev.ge.27.120193.002253
Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM (2002) Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295(5556):865–868. doi:10.1126/science.1067389
Kieran D, Kalmar B, Dick JR, Riddoch-Contreras J, Burnstock G, Greensmith L (2004) Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat Med 10(4):402–405. doi:10.1038/nm1021
Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ (2004) Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol 11(12):1215–1222. doi:10.1038/nsmb860
Goldberg A (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426(6968):895–904. doi:10.1038/nature02263
Pickart C (2004) Back to the future with ubiquitin. Cell 116(2):181–271
Baumeister W, Walz J, Zühl F, Seemüller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92(3):367–447
Powers E, Morimoto R, Dillin A, Kelly J, Balch W (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78:959–1050. doi:10.1146/annurev.biochem.052308.114844
Grune T, Jung T, Merker K, Davies K (2004) Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol 36(12):2519–2549. doi:10.1016/j.biocel.2004.04.020
Paul S (2008) Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays 30(11–12):1172–1256. doi:10.1002/bies.20852
Dobson CM (2003) Protein folding and misfolding. Nature 426(6968):884–890. doi:10.1038/nature02261
Harjes P, Wanker E (2003) The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci 28(8):425–458
Sullivan E, Weirich C, Guyon J, Sif S, Kingston R (2001) Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol 21(17):5826–5863
Yamanaka T, Miyazaki H, Oyama F, Kurosawa M, Washizu C, Doi H, Nukina N (2008) Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J 27(6):827–866. doi:10.1038/emboj.2008.23
Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144(1):67–78. doi:10.1016/j.cell.2010.11.050
Kopito R (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10(12):524–554
Olzmann J, Li L, Chin L (2008) Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem 15(1):47–107
Gentry M, Worby C, Dixon J (2005) Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A 102(24):8501–8507. doi:10.1073/pnas.0503285102
Rao SN, Maity R, Sharma J, Dey P, Shankar SK, Satishchandra P, Jana NR (2010) Sequestration of chaperones and proteasome into Lafora bodies and proteasomal dysfunction induced by Lafora disease-associated mutations of malin. Hum Mol Genet 19(23):4726–4734. doi:10.1093/hmg/ddq407
Singh S, Satishchandra P, Shankar SK, Ganesh S (2008) Lafora disease in the Indian population: EPM2A and NHLRC1 gene mutations and their impact on subcellular localization of laforin and malin. Hum Mutat 29(6):E1–12. doi:10.1002/humu.20737
Kahle PJ, Haass C (2004) How does parkin ligate ubiquitin to Parkinson’s disease? EMBO Rep 5(7):681–685. doi:10.1038/sj.embor.74001887400188
Lenartowski R, Gumowski K, Goc A (2008) The multifunctionality of CHIP protein in the protein quality-control system. Postepy Hig Med Dosw (Online) 62:297–308
Chhangani D, Joshi AP, Mishra A (2012) E3 ubiquitin ligases in protein quality control mechanism. Mol Neurobiol 45(3):571–585. doi:10.1007/s12035-012-8273-x
Fu L, Gao Y-S, Sztul E (2005) Transcriptional repression and cell death induced by nuclear aggregates of non-polyglutamine protein. Neurobiol Dis 20(3):656–721. doi:10.1016/j.nbd.2005.05.015
Mishra A, Dikshit P, Purkayastha S, Sharma J, Nukina N, Jana NR (2008) E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem 283(12):7648–7656. doi:10.1074/jbc.M706620200
Mishra A, Godavarthi SK, Jana NR (2009) UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis 36(1):26–34. doi:10.1016/j.nbd.2009.06.010
Mishra A, Jana NR (2008) Regulation of turnover of tumor suppressor p53 and cell growth by E6-AP, a ubiquitin protein ligase mutated in Angelman mental retardation syndrome. Cell Mol Life Sci 65(4):656–666. doi:10.1007/s00018-007-7476-1
Johnston J, Ward C, Kopito R (1998) Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143(7):1883–1981
Wigley W, Fabunmi R, Lee M, Marino C, Muallem S, DeMartino G, Thomas P (1999) Dynamic association of proteasomal machinery with the centrosome. J Cell Biol 145(3):481–571
Mitsui K, Nakayama H, Akagi T, Nekooki M, Ohtawa K, Takio K, Hashikawa T, Nukina N (2002) Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J Neurosci 22(21):9267–9344
Li JY, Englund E, Widner H, Rehncrona S, Björklund A, Lindvall O, Brundin P (2010) Characterization of Lewy body pathology in 12- and 16-year-old intrastriatal mesencephalic grafts surviving in a patient with Parkinson’s disease. Mov Disord 25(8):1091–1097. doi:10.1002/mds.23012
Bazan NG, Eady TN, Khoutorova L, Atkins KD, Hong S, Lu Y, Zhang C, Jun B, Obenaus A, Fredman G, Zhu M, Winkler JW, Petasis NA, Serhan CN, Belayev L (2012) Novel aspirin-triggered neuroprotectin D1 attenuates cerebral ischemic injury after experimental stroke. Exp Neurol 236(1):122–130. doi:10.1016/j.expneurol.2012.04.007
Calandria JM, Mukherjee PK, de Rivero Vaccari JC, Zhu M, Petasis NA, Bazan NG (2012) Ataxin-1 poly(Q)-induced proteotoxic stress and apoptosis are attenuated in neural cells by docosahexaenoic acid-derived neuroprotectin D1. J Biol Chem 287(28):23726–23739. doi:10.1074/jbc.M111.287078
Schubert U, Antón L, Gibbs J, Norbury C, Yewdell J, Bennink J (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404(6679):770–774. doi:10.1038/35008096
Yewdell J, Antón L, Bennink J (1996) Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J Immunol 157(5):1823–1829
Berke S, Paulson H (2003) Protein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Curr Opin Genet Dev 13(3):253–314
Heck J, Cheung S, Hampton R (2010) Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A 107(3):1106–1117. doi:10.1073/pnas.0910591107
Matsuo Y, Kishimoto H, Tanae K, Kitamura K, Katayama S, Kawamukai M (2011) Nuclear protein quality is regulated by the ubiquitin-proteasome system through the activity of Ubc4 and San1 in fission yeast. J Biol Chem 286(15):13775–13865. doi:10.1074/jbc.M110.169953
Hampton R (2011) San1-mediated quality control: substrate recognition “sans” chaperones. Mol Cell 41(1):2–5. doi:10.1016/j.molcel.2010.12.022
Rosenbaum J, Fredrickson E, Oeser M, Garrett-Engele C, Locke M, Richardson L, Nelson Z, Hetrick E, Milac T, Gottschling D, Gardner R (2011) Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell 41(1):93–199. doi:10.1016/j.molcel.2010.12.004
Bashan A, Yonath A (2008) Correlating ribosome function with high-resolution structures. Trends Microbiol 16(7):326–361. doi:10.1016/j.tim.2008.05.001
Brandt F, Etchells S, Ortiz J, Elcock A, Hartl F, Baumeister W (2009) The native 3D organization of bacterial polysomes. Cell 136(2):261–332. doi:10.1016/j.cell.2008.11.016
Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E (2010) A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol 189(1):57–68. doi:10.1083/jcb.200910074
Kramer G, Boehringer D, Ban N, Bukau B (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16(6):589–686. doi:10.1038/nsmb.1614
Rauch T, Hundley H, Pfund C, Wegrzyn R, Walter W, Kramer G, Kim S-Y, Craig E, Deuerling E (2005) Dissecting functional similarities of ribosome-associated chaperones from Saccharomyces cerevisiae and Escherichia coli. Mol Microbiol 57(2):357–422. doi:10.1111/j.1365-2958.2005.04690.x
Bengtson MH, Joazeiro CA (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467(7314):470–473. doi:10.1038/nature09371
Chu J, Hong N, Masuda C, Jenkins B, Nelms K, Goodnow C, Glynne R, Wu H, Masliah E, Joazeiro C, Kay S (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci U S A 106(7):2097–2200. doi:10.1073/pnas.0812819106
Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies R (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol 11(10):3425–3464
Collart M (2003) Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313:1–17
Panasenko O, Collart M (2012) Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol Microbiol 83(3):640–693. doi:10.1111/j.1365-2958.2011.07957.x
Gao X, Hu H (2008) Quality control of the proteins associated with neurodegenerative diseases. Acta Biochim Biophys Sin (Shanghai) 40(7):612–618
Ellis J (1987) Proteins as molecular chaperones. Nature 328:378–387. doi:10.1038/328378a0
Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A (1997) Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem 272(14):9002–9010
Lee D, Sherman M, Goldberg A (1996) Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol 16(9):4773–4854
McClellan A, Frydman J (2001) Molecular chaperones and the art of recognizing a lost cause. Nat Cell Biol 3(2):E51–3. doi:10.1038/35055162
Plemper R, Böhmler S, Bordallo J, Sommer T, Wolf D (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388(6645):891–896. doi:10.1038/42276
Ballinger C, Connell P, Wu Y, Hu Z, Thompson L, Yin L, Patterson C (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 19(6):4535–4580
Murata S, Minami Y, Minami M, Chiba T, Tanaka K (2001) CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2(12):1133–1141. doi:10.1093/embo-reports/kve246
Ehrlich E, Wang T, Luo K, Xiao Z, Niewiadomska A, Martinez T, Xu W, Neckers L, Yu X-F (2009) Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc Natl Acad Sci U S A 106(48):20330–20335. doi:10.1073/pnas.0810571106
Mishra A, Godavarthi SK, Maheshwari M, Goswami A, Jana NR (2009) The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J Biol Chem 284(16):10537–10545. doi:10.1074/jbc.M806804200
Ying Z, Wang H, Fan H, Zhu X, Zhou J, Fei E, Wang G (2009) Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum Mol Genet 18(22):4268–4349. doi:10.1093/hmg/ddp380
Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K (2008) Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol Biol Cell 19(4):1328–1336. doi:10.1091/mbc.E07-06-0601
Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400(6745):687–693. doi:10.1038/23293
Guo X, Shen S, Song S, He S, Cui Y, Xing G, Wang J, Yin Y, Fan L, He F, Zhang L (2011) The E3 ligase Smurf1 regulates Wolfram syndrome protein stability at the endoplasmic reticulum. J Biol Chem 286(20):18037–18047. doi:10.1074/jbc.M111.225615
Xu C, Bailly-Maitre B, Reed JC (2005) Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 115(10):2656–2664. doi:10.1172/JCI26373
Egger L, Madden DT, Rheme C, Rao RV, Bredesen DE (2007) Endoplasmic reticulum stress-induced cell death mediated by the proteasome. Cell Death Differ 14(6):1172–1180. doi:10.1038/sj.cdd.4402125
Roth W, Kermer P, Krajewska M, Welsh K, Davis S, Krajewski S, Reed JC (2003) Bifunctional apoptosis inhibitor (BAR) protects neurons from diverse cell death pathways. Cell Death Differ 10(10):1178–1187. doi:10.1038/sj.cdd.44012874401287
Deak PM, Wolf DH (2001) Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem 276(14):10663–10669. doi:10.1074/jbc.M008608200M008608200
Lerner M, Corcoran M, Cepeda D, Nielsen M, Zubarev R, Pontén F, Uhlén M, Hober S, Grandér D, Sangfelt O (2007) The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell 18(5):1670–1752. doi:10.1091/mbc.E06-03-0248
Maruyama Y, Yamada M, Takahashi K (2008) Ubiquitin ligase Kf-1 is involved in the endoplasmic reticulum-associated degradation pathway. Biochem Biophys Res Commun 374(4):737–741. doi:10.1016/j.bbrc.2008.07.126
Yasojima K, Tsujimura A, Mizuno T, Shigeyoshi Y, Inazawa J, Kikuno R, Kuma K, Ohkubo K, Hosokawa Y, Ibata Y, Abe T, Miyata T, Matsubara K, Nakajima K, Hashimoto-Gotoh T (1997) Cloning of human and mouse cDNAs encoding novel zinc finger proteins expressed in cerebellum and hippocampus. Biochem Biophys Res Commun 231(2):481–487
Kreft SG, Wang L, Hochstrasser M (2006) Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J Biol Chem 281(8):4646–4653. doi:10.1074/jbc.M512215200
Zavacki AM, Arrojo EDR, Freitas BC, Chung M, Harney JW, Egri P, Wittmann G, Fekete C, Gereben B, Bianco AC (2009) The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol 29(19):5339–5347. doi:10.1128/MCB.01498-08
Shah IM, Di Napoli M (2007) The ubiquitin-proteasome system and proteasome inhibitors in central nervous system diseases. Cardiovasc Hematol Disord Drug Targets 7(4):250–273
Kwon Y, Reiss Y, Fried V, Hershko A, Yoon J, Gonda D, Sangan P, Copeland N, Jenkins N, Varshavsky A (1998) The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci U S A 95(14):7898–8801
Zenker M, Mayerle J, Lerch M, Tagariello A, Zerres K, Durie P, Beier M, Hülskamp G, Guzman C, Rehder H, Beemer F, Hamel B, Vanlieferinghen P, Gershoni-Baruch R, Vieira M, Dumic M, Auslender R, Gil-da-Silva-Lopes V, Steinlicht S, Rauh M, Shalev S, Thiel C, Ekici A, Winterpacht A, Kwon Y, Varshavsky A, Reis A (2005) Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson–Blizzard syndrome). Nat Genet 37(12):1345–1395. doi:10.1038/ng1681
Johnson E, Ma P, Ota I, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270(29):17442–17498
Koegl M, Hoppe T, Schlenker S, Ulrich H, Mayer T, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96(5):635–679
Hwang CS, Shemorry A, Varshavsky A (2009) Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc Natl Acad Sci U S A 106(7):2142–2147. doi:10.1073/pnas.0812316106
Hwang CS, Shemorry A, Auerbach D, Varshavsky A (2010) The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol 12(12):1177–1262. doi:10.1038/ncb2121
Nillegoda N, Theodoraki M, Mandal A, Mayo K, Ren H, Sultana R, Wu K, Johnson J, Cyr D, Caplan A (2010) Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell 21(13):2102–2118. doi:10.1091/mbc.E10-02-0098
Eisele F, Wolf DH (2008) Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett 582(30):4143–4146. doi:10.1016/j.febslet.2008.11.015
Varshavsky A Recent studies of the ubiquitin system and the N-end rule pathway. Harvey Lect 96:93-209
Varshavsky A (2005) Regulated protein degradation. Trends Biochem Sci 30(6):283–289. doi:10.1016/j.tibs.2005.04.005
Vashist S, Kim W, Belden W, Spear E, Barlowe C, Ng D (2001) Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol 155(3):355–423. doi:10.1083/jcb.200106123
Wilhovsky S, Gardner R, Hampton R (2000) HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell 11(5):1697–2405
Ross C, Poirier M (2004) Protein aggregation and neurodegenerative disease. Nat Med 10(Suppl):S10–7. doi:10.1038/nm1066
Nakamura T, Lipton S (2009) Cell death: protein misfolding and neurodegenerative diseases. Apoptosis 14(4):455–523. doi:10.1007/s10495-008-0301-y
Kubota H (2009) Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem 146(5):609–625. doi:10.1093/jb/mvp139
Kishino T, Lalande M, Wagstaff J (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15(1):70–73. doi:10.1038/ng0197-70
Chan E, Young E, Ianzano L, Munteanu I, Zhao X, Christopoulos C, Avanzini G, Elia M, Ackerley C, Jovic N, Bohlega S, Andermann E, Rouleau G, Delgado-Escueta A, Minassian B, Scherer S (2003) Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 35(2):125–132. doi:10.1038/ng1238
Mittal S, Dubey D, Yamakawa K, Ganesh S (2007) Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum Mol Genet 16(7):753–815. doi:10.1093/hmg/ddm006
Junn E, Lee S, Suhr U, Mouradian M (2002) Parkin accumulation in aggresomes due to proteasome impairment. J Biol Chem 277(49):47870–47877. doi:10.1074/jbc.M203159200
Muqit M, Davidson S, Payne Smith M, MacCormac L, Kahns S, Jensen P, Wood N, Latchman D (2004) Parkin is recruited into aggresomes in a stress-specific manner: over-expression of parkin reduces aggresome formation but can be dissociated from parkin’s effect on neuronal survival. Hum Mol Genet 13(1):117–152. doi:10.1093/hmg/ddh012
Zhao J, Ren Y, Jiang Q, Feng J (2003) Parkin is recruited to the centrosome in response to inhibition of proteasomes. J Cell Sci 116(Pt 19):4011–4020. doi:10.1242/jcs.00700
Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286(5446):1882–1888
Fiedler K, Simons K (1995) The role of N-glycans in the secretory pathway. Cell 81(3):309–312
Helenius A, Aebi M (2001) Intracellular functions of N-linked glycans. Science 291(5512):2364–2369
Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K, Tai T (2002) E3 ubiquitin ligase that recognizes sugar chains. Nature 418(6896):438–442. doi:10.1038/nature00890
Thalmann R, Henzl MT, Thalmann I (1997) Specific proteins of the organ of Corti. Acta Otolaryngol 117(2):265–268
Nelson RF, Glenn KA, Zhang Y, Wen H, Knutson T, Gouvion CM, Robinson BK, Zhou Z, Yang B, Smith RJ, Paulson HL (2007) Selective cochlear degeneration in mice lacking the F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase subunit. J Neurosci 27(19):5163–5171. doi:10.1523/JNEUROSCI.0206-07.2007
Yoshida Y, Tokunaga F, Chiba T, Iwai K, Tanaka K, Tai T (2003) Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J Biol Chem 278(44):43877–43884. doi:10.1074/jbc.M304157200
Yoshida Y, Adachi E, Fukiya K, Iwai K, Tanaka K (2005) Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep 6(3):239–244. doi:10.1038/sj.embor.7400351
Groisman B, Avezov E, Lederkremer GZ (2006) The E3 ubiquitin ligases HRD1 and SCFFbs2 recognize the protein moiety and sugar chains, respectively, of an ER-associated degradation substrate. Israel Journal of Chemistry 46(2):189–196
Acknowledgments
This work was supported by the Department of Biotechnology, Government of India. AM was supported by Ramalinganswami Fellowship from the Department of Biotechnology, Government of India. The authors would like to thank Mr. Bharat Pareek for his support and management during manuscript preparation.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chhangani, D., Jana, N.R. & Mishra, A. Misfolded Proteins Recognition Strategies of E3 Ubiquitin Ligases and Neurodegenerative Diseases. Mol Neurobiol 47, 302–312 (2013). https://doi.org/10.1007/s12035-012-8351-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-012-8351-0