Abstract
Type 1 diabetes (T1D) is a complex autoimmune disease, and first stages of the disease typically develop early in life. Genetic as well as environmental factors are thought to contribute to the risk of developing autoimmunity against pancreatic beta cells. Several environmental factors, such as breastfeeding or early introduction of solid food, have been associated with increased risk for developing T1D. During the first years of life, the gut microbial community is shaped by the environment, in particular by dietary factors. Moreover, the gut microbiome has been described for its role in shaping the immune system early in life and early data suggest associations between T1D risk and alterations in gut microbial communities. In this article, we discuss environmental factors influencing the colonization process of the gut microbial community. Furthermore, we review possible interactions between the microbiome and the host that might contribute to the risk of developing T1D.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Krischer JP, Lynch KF, Schatz DA, et al. The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia. 2015;58(5):980–7. Interesting data from the TEDDY study, showing the incidence of autoantibody development over time.
Eringsmark Regnell S, Lernmark A. The environment and the origins of islet autoimmunity and Type 1 diabetes. Diabet Med. 2013;30(2):155–60.
Knip M, Simell O. Environmental triggers of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2(7):a007690.
Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41.
Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–90.
Sommer F, Backhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–38. Useful review article discussing the role of the gut microbiome in the development of the host immune system, metabolsim and physiology.
Brown CT, Davis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792.
Davis-Richardson AG, Ardissone AN, Dias R, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678.
Endesfelder D, zu Castell W, Ardissone A, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes. 2014;63(6):2006–14.
Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91.
Endesfelder D, Engel M, Davis-Richardson AG, et al. Towards a functional hypothesis relating anti-islet cell autoimmunity to the dietary impact on microbial communities and butyrate production. Microbiome. 2016; 4(1):17.
Kostic AD, Gevers D, Siljander H, et al. The dynamics of the human infant gut microbiome in development and in progression toward Type 1 diabetes. Cell Host Microbe. 2015;17(2):260–73.
Murri M, Leiva I, Gomez-Zumaquero JM, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case–control study. BMC Med. 2013;11:46.
Mejia-Leon ME, Petrosino JF, Ajami NJ, et al. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4:3814.
de Goffau MC, Luopajarvi K, Knip M, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62(4):1238–44.
Alkanani AK, Hara N, Gottlieb PA, et al. Alterations in intestinal microbiota correlate with susceptibility to Type 1 diabetes. Diabetes. 2015;64(10):3510–20.
Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98(2):229–38.
Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4578–85.
Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177.
Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65.
Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–5.
Martin R, Langa S, Reviriego C, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143(6):754–8.
Kawada M, Okuzumi K, Hitomi S, et al. Transmission of Staphylococcus aureus between healthy, lactating mothers and their infants by breastfeeding. J Hum Lact. 2003;19(4):411–7.
Gronlund MM, Lehtonen OP, Eerola E, et al. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28(1):19–25.
Lara-Villoslada F, Olivares M, Sierra S, et al. Beneficial effects of probiotic bacteria isolated from breast milk. Br J Nutr. 2007;98 Suppl 1:S96–100.
Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–7.
Turroni F, Peano C, Pass DA, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7(5):e36957.
Bergstrom A, Skov TH, Bahl MI, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80(9):2889–900.
Marques TM, Wall R, Ross RP, et al. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol. 2010;21(2):149–56.
Boehm G, Moro G. Structural and functional aspects of prebiotics used in infant nutrition. J Nutr. 2008;138(9):1818S–28S.
Oozeer R, van Limpt K, Ludwig T, et al. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am J Clin Nutr. 2013;98(2):561S–71S.
Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–21.
Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–35. This is an Important review article that discusses several factors how differences in glycan availability influence the gut microbial community.
Marcobal A, Barboza M, Sonnenburg ED, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10(5):507–14.
Miwa M, Horimoto T, Kiyohara M, et al. Cooperation of beta-galactosidase and beta-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20(11):1402–9.
LoCascio RG, Desai P, Sela DA, et al. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76(22):7373–81.
Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149(1):58–64.
Schwab C, Ganzle M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol Lett. 2011;315(2):141–8.
Martinez RC, Aynaou AE, Albrecht S, et al. In vitro evaluation of gastrointestinal survival of Lactobacillus amylovorus DSM 16698 alone and combined with galactooligosaccharides, milk and/or Bifidobacterium animalis subsp. lactis Bb-12. Int J Food Microbiol. 2011;149(2):152–8.
van Passel MW, Kant R, Zoetendal EG, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. 2011;6(3):e16876.
Goodman AL, Kallstrom G, Faith JJ, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108(15):6252–7.
Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14.
Faith JJ, McNulty NP, Rey FE, et al. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333(6038):101–4.
Englyst HN, Kingman SM, Cummings JH. Classification and measurement of nutritionally important starch fractions. Eur J Clin Nutr. 1992;46 Suppl 2:S33–50.
Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13(11):790–801.
Martinez FA, Balciunas EM, Converti A, et al. Bacteriocin production by Bifidobacterium spp A review. Biotechnol Adv. 2013;31(4):482–8.
De Vuyst L, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol. 2011;149(1):73–80.
Chang PV, Hao L, Offermanns S, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111(6):2247–52.
Furusawa Y, Obata Y, Fukuda S. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–50. This important study shows that butyrate production by commensal microbiota induces the differentiation of regulatory T cells in the colonic lamina propria of mice.
Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–5.
Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–73.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23.
Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105(39):15064–9.
Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4659–65.
Johansson, M.E., H.E. Jakobsson, J. Holmen-Larsson, et al., Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe, 2015
Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14(7):660–7.
Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–68.
Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105(52):20858–63.
Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–8.
Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303(5664):1662–5.
Kawamoto S, Tran TH, Maruya M, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336(6080):485–9.
Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–27.
Lee YK, Menezes JS, Umesaki Y, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4615–22.
Arrieta MC, Finlay BB. The commensal microbiota drives immune homeostasis. Front Immunol. 2012;3:33.
Ivanov II, Atarashi K, Mane N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98.
Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–89.
Atarashi K, Nishimura J, Shima T, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455(7214):808–12.
Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–9.
Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41.
Geuking MB, Cahenzli J, Lawson MA, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806.
Hoeppli RE, Wu D, Cook L, et al. The environment of regulatory T cell biology: cytokines, metabolites, and the microbiome. Front Immunol. 2015;6:61.
Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–4.
Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14(7):676–84.
An D, Oh SF, Olszak T, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156(1–2):123–33.
Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–19.
Finnie IA, Dwarakanath AD, Taylor BA, et al. Colonic mucin synthesis is increased by sodium butyrate. Gut. 1995;36(1):93–9.
Shimotoyodome A, Meguro S, Hase T, et al. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp Biochem Physiol A Mol Integr Physiol. 2000;125(4):525–31.
Pryde SE, Duncan SH, Hold GL, et al. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217(2):133–9.
Duncan SH, Holtrop G, Lobley GE, et al. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91(6):915–23.
Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66(4):1654–61.
Louis P, Duncan SH, McCrae SI, et al. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186(7):2099–106.
Flint HJ, Duncan SH, Scott KP, et al. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9(5):1101–11.
Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett. 2006;254(1):116–22.
Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–72.
Dar SA, Kleerebezem R, Stams AJ, et al. Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio. Appl Microbiol Biotechnol. 2008;78(6):1045–55.
Willis CL, Cummings JH, Neale G, et al. In vitro effects of mucin fermentation on the growth of human colonic sulphate-reducing bacteria. Anaerobe. 1996;2(2):117–22.
KEGG Database, http://www.genome.jp/kegg-bin/show_pathway?amu00920. Accessed 26 November 2015.
Vaarala O. Is the origin of type 1 diabetes in the gut? Immunol Cell Biol. 2012;90(3):271–6.
Vaarala O. Leaking gut in type 1 diabetes. Curr Opin Gastroenterol. 2008;24(6):701–6.
Kuitunen M, Saukkonen T, Ilonen J, et al. Intestinal permeability to mannitol and lactulose in children with type 1 diabetes with the HLA-DQB1*02 allele. Autoimmunity. 2002;35(5):365–8.
Sapone A, de Magistris L, Pietzak M, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. 2006;55(5):1443–9.
Secondulfo M, Iafusco D, Carratu R, et al. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis. 2004;36(1):35–45.
Bosi E, Molteni L, Radaelli MG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49(12):2824–7.
Mooradian AD, Morley JE, Levine AS, et al. Abnormal intestinal permeability to sugars in diabetes mellitus. Diabetologia. 1986;29(4):221–4.
Rook GA, Brunet LR. Microbes, immunoregulation, and the gut. Gut. 2005;54(3):317–20.
Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–62.
Bach JF, Chatenoud L. The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harb Perspect Med. 2012;2(2):a007799.
Virtanen SM, Kenward MG, Erkkola M, et al. Age at introduction of new foods and advanced beta cell autoimmunity in young children with HLA-conferred susceptibility to type 1 diabetes. Diabetologia. 2006;49(7):1512–21.
Cardwell CR, Stene LC, Ludvigsson J, et al. Breast-feeding and childhood-onset type 1 diabetes: a pooled analysis of individual participant data from 43 observational studies. Diabetes Care. 2012;35(11):2215–25.
Ziegler AG, Schmid S, Huber D, et al. Early infant feeding and risk of developing type 1 diabetes-associated autoantibodies. JAMA. 2003;290(13):1721–8.
Chmiel R, Beyerlein A, Knopff A, et al. Early infant feeding and risk of developing islet autoimmunity and type 1 diabetes. Acta Diabetol. 2015;52(3):621–4.
Norris JM, Barriga K, Klingensmith G, et al. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA. 2003;290(13):1713–20.
Aronsson CA, Lee HS, Liu E, et al. Age at gluten introduction and risk of celiac disease. Pediatrics. 2015;135(2):239–45.
Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7.
Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–62.
Semenkovich CF, Danska J, Darsow T, et al. American Diabetes Association and JDRF Research Symposium: diabetes and the microbiome. Diabetes. 2015;64(12):3967–77.
Karvonen M, Viik-Kajander M, Moltchanova E, et al. Incidence of childhood type 1 diabetes worldwide. Diabetes Mondiale (DiaMond) Project Group. Diabetes Care. 2000;23(10):1516–26.
Kemppainen KM, Ardissone AN, Davis-Richardson AG, et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38(2):329–32.
Cardwell CR, Stene LC, Joner G, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51(5):726–35.
Bonifacio E, Warncke K, Winkler C, et al. Cesarean section and interferon-induced helicase gene polymorphisms combine to increase childhood type 1 diabetes risk. Diabetes. 2011;60(12):3300–6.
Vehik K, Dabelea D. Why are C-section deliveries linked to childhood type 1 diabetes? Diabetes. 2012;61(1):36–7.
De Palma G, Capilla A, Nadal I, et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr Issues Mol Biol. 2010;12(1):1–10.
Rausch P, Rehman A, Kunzel S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108(47):19030–5.
Yang P, Li HL, Wang CY. FUT2 nonfunctional variant: a “missing link” between genes and environment in type 1 diabetes? Diabetes. 2011;60(11):2685–7.
Group TS. The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann N Y Acad Sci. 2008;1150:1–13.
Mills S, Shanahan F, Stanton C, et al. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4(1):4–16.
De Paepe M, Leclerc M, Tinsley CR, et al. Bacteriophages: an underestimated role in human and animal health? Front Cell Infect Microbiol. 2014;4:39.
Acknowledgments
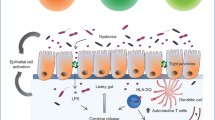
David Endesfelder and Wolfgang zu Castell report grants from the Juvenile Diabetes Research Foundation. We thank Ms. Sandra Mayer for the production of Fig. 1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
David Endesfelder, Marion Engel, and Wolfgang zu Castell declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Immunology and Transplantation
Rights and permissions
About this article
Cite this article
Endesfelder, D., Engel, M. & zu Castell, W. Gut Immunity and Type 1 Diabetes: a Mélange of Microbes, Diet, and Host Interactions?. Curr Diab Rep 16, 60 (2016). https://doi.org/10.1007/s11892-016-0753-3
Published:
DOI: https://doi.org/10.1007/s11892-016-0753-3