Abstract
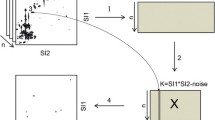
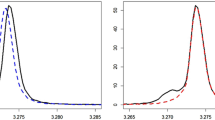
A modified Lorentzian distribution function is used to model peaks in two-dimensional (2D) 1H–13C heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectra. The model fit is used to determine accurate chemical shifts from genuine signals in complex metabolite mixtures such as blood. The algorithm can be used to extract features from a set of spectra from different samples for exploratory metabolomics. First a reference spectrum is created in which the peak intensities are given by the median value over all samples at each point in the 2D spectra so that 1H–13C correlations in any spectra are accounted for. The mathematical model provides a footprint for each peak in the reference spectrum, which can be used to bin the 1H–13C correlations in each HSQC spectrum. The binned intensities are then used as variables in multivariate analyses and those found to be discriminatory are rapidly identified by cross referencing the chemical shifts of the bins with a database of 13C and 1H chemical shift correlations from known metabolites.





Similar content being viewed by others
References
Ali, K., Maltese, F., Zyprian, E., Rex, M., Choi, Y. H., & Verpoorte, R. (2009). NMR metabolic fingerprinting based identification of grapevine metabolites associated with downy mildew resistance. Journal of Agricultural and Food Chemistr, 57, 9599–9606.
Alsberg, B. K., Goodacre, R., Rowland, J. J., & Kell, D. B. (1997). Classification of pyrolysis mass spectra by fuzzy multivariate rule induction-comparison with regression, K-nearest neighbour, neural and decision-tree methods. Analytica Chimica Acta, 348, 389–407.
Bailey, N. J. C., Oven, M., Holmes, E., Nicholson, J. K., & Zenk, M. H. (2003). Metabolomic analysis of the consequences of cadmium exposure in Silene cucubalus cell cultures via 1H NMR spectroscopy and chemometrics. Phytochemistry, 62, 851–858.
Bollard, M. E., Stanley, E. G., Lindon, J. C., Nicholson, J. K., & Holmes, E. (2005). NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR in Biomedicine, 18, 143–162.
Charlton, A., Allnutt, T., Holmes, S., Chisholm, J., Bean, S., Ellis, N., et al. (2004). NMR profiling of transgenic peas. Plant Biotechnology Journal, 2, 27–35.
Charlton, A. J., Donarski, J. A., Harrison, M., Jones, S. A., Godward, J., Oehlschlager, S., et al. (2008). Responses of the pea (Pisum sativum L.) leaf metabolome to drought stress assessed by nuclear magnetic resonance spectroscopy. Metabolomics, 4, 312–327.
Choi, H.-K., Yoon, J.-H., Kim, Y.-S., & Kwon, D. Y. (2007). Metabolomic profiling of Cheonggukjang during fermentation by 1H NMR spectrometry and principal components analysis. Process Biochemistry, 42, 263–266.
Cubbon, S., Bradbury, T., Wilson, J., & Thomas-Oates, J. (2007). Hydrophilic interaction chromatography for mass spectrometric metabonomic studies of urine. Analytical Chemistry, 79, 8911–8918.
Davis, R. A., Charlton, A. J., Godward, J., Jones, S. A., Harrison, M., & Wilson, J. C. (2007). Adaptive binning: An improved binning method for metabolomics data using the undecimated wavelet transform. Chemometrics and Intelligent Laboratory Systems, 85, 144–154.
Davis, R. A., Charlton, A. J., Oehlschlager, S., & Wilson, J. (2006). A novel feature selection method for genetic programming using 1H NMR data. Chemometrics and Intelligent Laboratory Systems, 81, 50–59.
Day, I. J., Mitchell, J. C., Snowden, M. J., & Davis, A. L. (2008). Investigation of the potential of the dissolution dynamic nuclear polarization method for general sensitivity enhancement in small-molecule NMR spectroscopy. Applied Magnetic Resonance, 34, 453–460.
De Meyer, T., Sinnaeve, D., Van Gasse, B., Tsiporkova, E., Rietzschel, E. R., De Buyzere, M. L., et al. (2008). NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Analytical Chemistry, 80, 3783–3790.
Forshed, J., Schuppe-Koistinen, I., & Jacobsson, S. P. (2003). Peak alignment of NMR signals by means of a genetic algorithm. Analytica Chimica Acta, 487, 189–199.
Frydman, L., & Blazina, D. (2007). Ultrafast two-dimensional nuclear magnetic resonance spectroscopy of hyperpolarized solutions. Nature Physics, 3, 415–419.
Giraudeau, P., Shrot, Y., & Frydman, L. (2009). Multiple ultrafast, broadband 2D NMR spectra of hyperpolarized natural products. Journal of the American Chemical Society, 131, 13902–13903.
Goodacre, R., Shann, B., Gilbert, R. J., Timmins, É. M., McGovern, A. C., Alsberg, B. K., et al. (2000). The detection of the dipicolinic acid biomarker in Bacillus spores using Curie-point pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. Analytical Chemistry, 72, 119–127.
Griffin, J. L., Williams, H. J., Sang, E., Clarke, K., Rae, C., & Nicholson, J. K. (2001). Metabolic profiling of genetic disorders: A multitissue 1H nuclear magnetic resonance spectroscopic and pattern recognition study into dystrophic tissue. Analytical Biochemistry, 293, 16–21.
Gronwald, W., Klein, M. S., Kaspar, H., Fagerer, S. R., Nürnberger, N., Dettmer, K., et al. (2008). Urinary metabolite quantification employing 2D NMR spectroscopy. Analytical Chemistry, 80, 9288–9297.
Gullberg, J., Jonsson, P., Nordstrom, A., Sjostrom, M., & Moritz, T. (2004). Design of experiments: An efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Analytical Biochemistry, 331, 283–295.
Hall, R., Beale, M., Fiehn, O., Hardy, N., Sumner, L., & Bino, R. (2002). Plant metabolomics: The missing link in functional genomics strategies. The Plant Cell, 14, 1437–1440.
Hirakawa, K., Koike, K., Uekusa, K., Nihira, M., Yuta, K., & Ohno, Y. (2009). Experimental estimation of postmortem interval using multivariate analysis of proton NMR metabolomic data. Legal Medicine, 11(Supplement 1), S282–S285.
Holmes, E., Nicholson, J. K., Nicholls, A. W., Lindon, J. C., Connor, S. C., Polley, S., et al. (1998). The identification of novel biomarkers of renal toxicity using automatic data reduction techniques and PCA of proton NMR spectra of urine. Chemometrics and Intelligent Laboratory Systems, 44, 245–255.
Hotelling, H. (1933). Analysis of a complex of statistical variables into principal components. Journal of Educational Psychology, 24(417–441), 498–520.
Jankevics, A., Liepinsh, E., Liepinsh, E., Vilskersts, R., Grinberga, S., Pugovics, O., et al. (2009). Metabolomic studies of experimental diabetic urine samples by 1H NMR spectroscopy and LC/MS method. Chemometrics and Intelligent Laboratory Systems, 97, 11–17.
Kaczmarek, K., Walczak, B., de Jong, S., & Vandeginste, B. G. M. (2002). Feature based fuzzy matching of 2D gel electrophoresis images. Journal of Chemical Information and Computer Sciences, 42, 1293–1305.
Koskela, H., Heikkilä, O., Kilpeläinen, I., & Heikkinen, S. (2010). Quantitative two-dimensional HSQC experiment for high magnetic field NMR spectrometers. Journal of Magnetic Resonance, 202, 24–33.
Lewis, I. A., Schommer, S. C., Hodis, B., Robb, K. A., Tonelli, M., Westler, W. M., et al. (2007). Method for determining molar concentrations of metabolites in complex solutions from two-dimensional 1H–13C NMR spectra. Analytical Chemistry, 79, 9385–9390.
Lewis, I. A., Schommer, S. C., & Markley, J. L. (2009). rNMR: Open source software for identifying and quantifying metabolites in NMR spectra. Magnetic Resonance in Chemistry, 47(S1), S123–S126.
Mehlkopf, A. F., Korbee, D., Tiggelman, T. A., & Freeman, R. (1984). Sources of t1 noise in two-dimensional NMR. Journal of Magnetic Resonance, 58, 315–323.
Mierisova, S., & Ala-korpela, M. (2001). MR spectroscopy quantitation: A review of frequency domain methods. NMR in Biomedicine, 14, 247–259.
Poulding, S., Charlton, A. J., Donarski, J., & Wilson, J. C. (2007). Removal of t1 noise from metabolomic 2D 1H–13C HSQC NMR spectra by correlated trace denoising. Journal of Magnetic Resonance, 189, 190–199.
Schleucher, J., Schwendinger, M., Sattler, M., Schmidt, P., Schedletzky, O., Glaser, S. J., et al. (1994). A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed-field gradients. Journal of Biomolecular NMR, 4, 301–306.
Tiziani, S., Schwartz, S. J., & Vodovotz, Y. (2006). Profiling of carotenoids in tomato juice by one- and two-dimensional NMR. Journal of Agricultural and Food Chemistry, 54, 6094–6100.
Turner, C. J., Connolly, P. J., & Stern, A. S. (1999). Artifacts in sensitivity-enhanced HSQC. Journal of Magnetic Resonance, 137, 281–284.
Ulrich, E. L., Akutsu, H., Doreleijers, J. F., Harano, Y., Ioannidis, Y. E., Lin, J., et al. (2008). BioMagResBank. Nucleic Acids Research, 36, D402–D408.
Ward, J. L., Harris, C., Lewis, J., & Beale, M. H. (2003). Assessment of 1H NMR spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry, 62, 949–957.
Wold, H. O. A. (1966). Estimation of principal components and related models by iterative least squares. In P. R. Krishnaiah (Ed.), Multivariate analysis (pp. 391–420). NY: Academic Press.
Wood, N. J., Brannigan, J. A., Duckett, S. B., Heath, S. L., & Wagstaff, J. (2007). Detection of picomole amounts of biological substrates by para-hydrogen-enhanced NMR methods in conjunction with a suitable receptor complex. Journal of the American Chemical Society, 129, 11012–11013.
Xi, Y., de Ropp, J. S., Viant, M. R., Woodruff, D. L., & Yu, P. (2008). Improved identification of metabolites in complex mixtures using HSQC NMR spectroscopy. Analytica Chimica Acta, 614, 127–133.
Acknowledgements
The authors would like to thank the Defra seedcorn programme for funding this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McKenzie, J.S., Charlton, A.J., Donarski, J.A. et al. Peak fitting in 2D 1H–13C HSQC NMR spectra for metabolomic studies. Metabolomics 6, 574–582 (2010). https://doi.org/10.1007/s11306-010-0226-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-010-0226-7