Abstract
Overexpression of genes that modify gibberellin (GA) metabolism and signaling have been previously shown to produce trees with improved biomass production but highly disturbed development. To examine if more subtle types of genetic modification of GA could improve growth rate and modify tree architecture, we transformed a model poplar genotype (Populus tremula × P. alba) with eight genes, including two cisgenes (intact copies of native genes), four intragenes (modified copies of native genes), and two transgenes (from sexually incompatible species), and studied their effects under greenhouse and field conditions. In the greenhouse, four out of the eight tested genes produced a significant and often striking improvement of stem volume, and two constructs significantly modified the proportion of root or shoot biomass. Characterization of GA concentrations in the cisgenic population that had an additional copy of a poplar GA20-oxidase gene showed elevated concentrations of 13-hydroxylated GAs compared to wild-type poplars. In the field, we observed growth improvement for three of the six tested constructs, but it was significantly greater for only one of the constructs, a pRGL:GA20-oxidase intragene. The greenhouse and field responses were highly variable, possibly to due to cross-talk among the GA pathway and other stress response pathways, or due to interactions between the cisgenes and intragenes with highly similar endogenes. Our results indicate that extensive field trials, similar to those required for conventional breeding, will be critical to evaluating the value and pleiotropic effects of GA-modifying genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gibberellins (GAs) are a large class of phytohormones that play fundamental and diverse roles in the regulation of plant growth and development (Olszewski et al. 2002; Achard and Genschik 2009; Hedden and Thomas 2012). Modification of GA levels in plants has been widely used in agriculture, for example to reduce stem elongation and subsequent lodging in cereals as part of the green revolution, to stimulate fruit growth in seedless grapes, to delay fruit senescence in oranges and lemons, or to increase stem elongation in sugarcane (reviewed in Hedden and Phillips 2000a, 2000b). The molecular biology and biochemistry of GA have been extensively reviewed (e.g., Busov et al. 2008; Yamaguchi 2008; Sponsel and Hedden 2010). Although more than 100 GAs have been identified in higher plants, fungi, and bacteria, only two are commonly biologically active per se within plants, while the others are generally biosynthetic precursors or catabolites. Thus, GA1 from the predominant early 13-hydroxylation pathway, and GA4 from the non-13-hydroxylation pathway, are the probable bioeffectors (Fig. 1; Rood and Hedden 1994; Olszewski et al. 2002). Their formation is catalyzed by GA20ox and GA3ox, and they are inactivated by GA2ox, and all of these enzymes are 2-oxoglutarate dependent dioxygenases (2-ODDs) and are subject to feedback regulation (reviewed by Yamaguchi and Kamiya 2000; Gallego-Giraldo et al. 2008; Hedden and Thomas 2012). The GA20ox and GA3ox genes are regulated by negative feedback as high levels of bioactive GAs lead to reduced expression of these genes. The GA2ox genes, on the other hand, are regulated by positive feedback, as high levels of bioactive GAs lead to increased expression of these genes, thereby promoting GA inactivation. The feedback regulations of these 2-ODD encoding genes play a central role for maintaining GA homeostasis.
Schematic diagram of the parallel gibberellin (GA) biosynthetic pathways in higher plants. The non-13-hydroxylation pathway (top) and early 13-hydroxylation pathway (bottom), leading to the probable bioeffectors, GA4 and especially GA1. The relevant enzymes are indicated and the GAs that were analyzed in our study are in bold and include the final non-13-hydroxylated GAs and the more abundant 13-hydroxylated GAs
GA20ox is involved in the catalysis of the penultimate steps in the biosynthesis of active GAs, and transgenic modification of its levels has been extensively studied in plants. GA20ox overexpression has been used to successfully modify plant stature in Arabidopsis thaliana (Arabidopsis) (AtGA20ox1; PdGA20ox1), Solanum tuberosum (StGA20ox1), tobacco (NtGA20ox1), hybrid aspen (PtGA20ox1; PdGA20ox1), Carrizo citrange (CcGA20ox1) ( reviewed by Bhattacharya et al. 2009; Jeon et al. 2015), tomato (CcGA20ox1) (García-Hurtado et al. 2012), rice (OsGA20ox3; Qin et al. 2013), and Tricyrtis sp. ‘Shinonome’ (TfGA20ox2; Otani et al. 2014). Regulators of GA action have also been used to modify plant development. Negative regulators like repressor of gal-3 (RGA), GA-insensitive (GAI), and repressor of gal-3 like (RGL) proteins from Arabidopsis are several well-studied DELLA domain proteins. In response to GA, these DELLA proteins rapidly degrade and repress GA action (Davière and Achard 2013; Claeys et al. 2014). Dominant mutations of DELLA proteins (e.g., rga, gai, and rgl1) have been shown to cause dwarfism in Arabidopsis, maize, wheat, barley, and Populus (Olszewski et al. 2002; Wen and Chang 2002; Busov et al. 2006). Another negative regulator is SPINDLY (SPY), which is an O-linked N-acetylglucosamine transferase that can interact with DELLA proteins (Claeys et al. 2014) and has been shown to inhibit GA signaling in Arabidopsis (Silverstone et al. 2007) and Petunia (Izhaki et al. 2001). Modification of SPY mRNA levels led to altered plant architecture, including reduced hypocotyl or stem elongation rate (Izhaki et al. 2001; Silverstone et al. 2007). The positive regulator photoperiod responsive 1 (PHOR1) was first identified in potato by Amador et al. (2001). PHOR1-overexpressing plants grew faster, while plants with inhibited PHOR1 expression presented a semi-dwarf phenotype and increased levels of endogenous GAs (reviewed by Olszewski et al. 2002).
Large modifications of tree form and architecture have been proposed as important steps in modifying trees for use in bioenergy plantations (Ragauskas et al. 2006; Mittal and Decker 2013). Key goals include rapid growth rate, reduced stature, and increased allocation to stems in relation to other woody tissues. Because GAs have prominent roles in the control of many aspects of vegetative development in plants, they are logical candidates as tools for achieving these modifications. Eriksson et al. (2000), Han et al. (2010) and Jeon et al. (2015) reported modified properties and improved biomass of poplars by transgenic or cisgenic modification of GA levels. Eriksson et al. (2000) overexpressed a modified GA20ox gene (lacking an upstream ATG codon) from Arabidopsis. The transgenic plants showed higher levels of C19 GAs (GA20, GA1, and GA8 from the 13-hydroxylation pathway, and GA9, GA4, and GA34 from the non-13-hydroxylation; Fig. 1), faster growth in height and diameter, and greater shoot vs. root biomass. Similarly, Jeon et al. (2015) overexpressed a Pinus densiflora GA20ox (PdGA20ox1) in poplars using a constitutive 35S promoter and a xylem-specific promoter respectively. In both cases, they observed an up to three-fold increase in stem biomass, but noticeable reduction in leaf area (20 % to 40 %) and root biomass (up to 80 %) in transgenic plants. In contrast, the poplars produced by Han et al. (2010) were transformed with five different cisgenes, including GA20ox7, GA2ox2, GAI1, RGL 1–1, and RGL 1–2. The GA20ox7 gene conferred an early growth advantage to the plants, but the effects diminished over time in the greenhouse trials. Other tested genes, in general, had a retarding effect on plant growth. These studies suggest that recombinant DNA (rDNA) modification of GA levels can improve tree growth and modify biomass allocation, but substantially more refinement of genetic constructs and phenotypic evaluation are needed before their potential value for application can be assessed.
In this study, we investigated the effects of several kinds of GA-modifying cisgenes, intragenes, and transgenes on the growth rate and architecture of poplars grown in both greenhouse and field environments (Fig. 2). We report GA-modifying rDNA can indeed alter GA concentrations, promote growth rate, and alter tree architecture; however, we also report marked instability of cisgenic, intragenic, and transgenic effects that suggest much additional phenotypic and physiological evaluation will be needed to understand their mechanisms of phenotypic modification and possible value for tree biotechnology.
Materials and methods
Promoters and genes selected
Eight promoter × coding region combinations of GA pathway components (Table 1) were used in our study. Three genes, GA20ox (PtGA20ox7 and PtGA20ox2-2), PHOR1 (PtPHOR1-2), and SPY (AtSPY and HvSPY), and seven promoters, either native or constitutive, were employed. For the first six constructs in Table 1, the promoters and the genes were selected based on a whole-genome microarray expression study (Rodgers-Melnick et al. 2012) with the goal of improving stem growth or altering biomass allocation. For the last two constructs in the table, we used SPY genes to explore their effect in poplar, which had not been studied previously.
Promoter cloning and testing
To clone four novel promoters, i.e., sequences upstream of PtRGL1-1, PtGA2ox1, PtEXPB1 (β-expansin), and PtCESA1 (cellulose synthase), we isolated genomic DNA from young leaves of Populus trichocarpa Nisqually-1 using the DNeasy Plant Mini Kit (Qiagen). We then amplified the upstream parts of these genes (1.5 to 3 kb, including 5′-UTR regions) from 100 ng of template genomic DNA using Platinum Tag DNA Polymerase High Fidelity (Invitrogen). Next, we cloned the PCR products into the pCR4-TOPO TA cloning vector (Invitrogen, hereafter named TOPO4 vector) and sequence verified five to eight colonies. The DNA sequences of each isolated promoter are given in electronic supplementary appendix S1.
To verify activity and examine expression patterns of these novel promoters in poplars, we inserted each of the promoters in front of an intron-containing GUS reporter gene in a GUS PLUS pCambia binary vector. We also inserted the promoter region of PtGA20ox7 (hereafter named promoter GA20) in the same pCambia vector. We used these five constructs (namely pGA2:GUS, pRGL:GUS, pEXP:GUS, pCES:GUS, and pGA20:GUS) to transform poplars. For GUS assays, we collected nine different tissues (Fig. S1) and incubated them in GUS solution (1 mg/ml X-Gluc, 100 mM sodium phosphate (pH 7.0), 0.5 % (v/v)Triton X-100, and 2 % (v/v) N,N-dimethylformamide) for 24 h at 37 °C, and then cleared away chlorophyll by soaking in 70 % ethanol and 10 % bleach for at least 6 h. For visual assessment, we scored GUS intensity for each stained tissue on a 0 to 3 scale (Fig. S2). To roughly quantify promoter specificity, we also calculated several types of specificity indexes, composed of ratios of mean scores between the expected target tissues for the promoters based on microarray or GUS data and other tissues. The lower the average score in non-target tissues, the higher will be the specificity index value; specificity index value greater than 1 indicates preferential expression in target tissue.
We were unable to analyze promoter RGL using GUS analysis because we found that, after transformation, a sequence error had been introduced; the GUS gene fused to the promoter was missing a start codon. Therefore, to confirm the activity of promoter RGL in pRGL:GA20- transformed poplars (Table 1), we first performed semi-quantitative reverse transcription-PCR (semi-quantitative RT-PCR) on leaf samples and verified construct-specific primers. We then conducted quantitative reverse transcription-PCR (qRT-PCR) to compare mRNA levels of the PtGA20ox2-2 gene in leaves and shoot tips. In brief, total RNA was extracted from shoot tips and young leaves of six pRGL:GA20-transformed events and one pBART control vector (described below under construct assembly) transformed event. Two independent sample collections (within 1 week) and extractions were made for each of the intragenic events and the empty-vector control. DNaseI-treated RNA was used for first-strand cDNA synthesis using SuperScript III Reverse Transcriptase (Invitrogen) and oligo (dT)18 primers according to the manufacturer’s protocol. The same primers were used for semi-quantitative RT-PCR and qRT-PCR (Table S1). The forward primer used for amplifying PtGA20ox2-2 was located in the gene, and the reverse primer was in the junction region between the PtGA20ox2-2 gene and the NOS terminator in the pRGL:GA20 construct. Therefore, this primer set specifically amplified a 167-bp region (49 bp of the PtGA20ox2-2 gene and 118 bp in the junction area (hereafter named “GA20 intragene”), without amplifying the endogenous PtGA20ox2-2 gene in poplar genome. Poplar ACTIN2 (Phytozome v9.1 accession Potri.001G453600; Table S1) was used as a control reference gene. Semi-quantitative RT-PCR was performed with RNA from leaf samples using 35 cycles of PCR amplification. qRT-PCR was performed with RNA from leaves and shoot tips respectively. For each sample type, we used one 96-well plate and included two biological pseudoreplicates and three technical replicates for each event. qRT-PCR was performed using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). PCR conditions were 95 °C for 10 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. qRT-PCR data was analyzed using comparative CT method.
Construct assembly
For the two cisgene constructs (pGA20 and pPHOR), we cloned a complete PCR product containing the entire upstream and downstream portions of the genes (PtGA20ox7 or PtPHOR1-2) into a TOPO4 vector. The fragment was then cloned into a pBART vector (based on pART27) (Gleave 1992) directly using NotI sites. The inserts were fully sequenced prior to cloning, and the plasmids were verified by sequencing of 5′ and 3′ ends regions after cloning. Details about the pBART vector and the cloning strategy of GA20 have been described earlier (Han et al. 2010).
To clone the coding region (PtGA20ox2-2) in the four intragene constructs (pRGL:GA20, pGA2:GA20, pEXP:GA20, and pCES:GA20), we first PCR-amplified the PtGA20ox2-2 transcript from shoot tips into a TOPO4 vector (forming pGA20TOPO4). We then ligated the 1189-bp EcoRI fragment from pGA20TOPO4 containing GA20ox2-2 into the EcoRI site of pCAMBIA 1390, in front of the NOS terminator. The transcript and the terminator were again PCR-amplified and the 1467-bp fragment was ligated into the PmeI site of the pBART vector (forming pGA20NOStBART).
Each novel promoter was then cloned into the pGA20NOStBART plasmid. Specifically, the RGL promoter and part of the RGL transcript were amplified together from genomic DNA and cloned into the TOPO4 vector (forming pRGLTOPO4); the RGL promoter was amplified from pRGLTOPO4 in the presence of Pfx polymerase and cloned into the PmeI site of pGA20NOStBART (forming pRGL:GA20NOStBART). Likewise, the GA2 promoter was first amplified from genomic DNA and then cloned into TOPO4 (forming pGA2TOPO4), from where the 1128-bp PmeI fragment was removed and cloned into pGA20NOStBART (forming pGA2:GA20NOStBART). Similarly, the EXP and the CES promoters were amplified from genomic DNA, cloned into TOPO4 vectors, sequence verified, and then cloned in front of the PtGA20ox2-2 coding sequence in pGA20NOStBART.
The two SPY transgene constructs (pATS and pHVS) were provided by Dr. Steve Swain (CSIRO Plant Industry, Australia). Cloning strategies were described in Swain et al. (2001) and Filardo et al. (2009). The barley HvSPY gene used in this study was originally named Hvspy-2 (Filardo et al. 2009); its sequence has deletions in exon 8.
Plant material, transformation and regeneration
The female hybrid clone 717-1B4 (Populus tremula × P. alba: INRA, France), and the Agrobacterium hypervirulent strain AGL1 (Chabaud et al. 2003) were used for plant transformation. In total, we transformed poplar plants with 14 different constructs, including five GUS constructs for promoter testing, eight GA-modifying constructs for greenhouse and field studies, and the pBART vector for control. Transformation and regeneration were essentially as described by Filichkin et al. (2006) except basta was used as the selection agent. Rooted plants were then PCR-verified for the presence of the GA gene of interest (primers in Table S1). Primers were designed to amplify partial border or junction fragments for cisgenic specific or promoter-PtGA20ox2-2 regions for intragenic specific. Wild-type control plants went through micropropagation under similar conditions to transformed plants. All experimental plants were propagated, acclimated to ambient humidity in gradually opened plastic bags over potted plants in soil, then transferred to 4-l pots containing SunGro Horticulture ‘Sunshine’ soil with Osmocote 14-14-14 slow-release fertilizer and moved to the greenhouse.
Greenhouse trial design and measurements
We performed a total of six greenhouse trials (from 2007 to 2009) and tested 236 rDNA modified events covering more than 1600 transformed plants (4–20 ramets/transformed event) (Table S2). The first greenhouse trial with the pATS and pHVS constructs included only wild-type control; the other five greenhouse trials included two types of controls, wild-type and empty-vector controls. The June 2008 and July 2008 trials employed a randomized block design (RBD), while the rest of the four trials had a completely randomized design (CRD). Three constructs (pPHOR, pRGL:GA20, and pGA2:GA20) were tested twice in greenhouse. Han et al. (2010) summarized the conditions maintained in the greenhouse. We supplemented natural daylight with artificial light from high pressure sodium lamps and fertilized the plants once every 3 months with Osmocote 20-20-20 slow release fertilizer. We recorded height and diameter (2 in. from the soil surface) every 2 weeks for 6 weeks, or weekly for 4 weeks. Volume index is the product of height and diameter2 and used as an index of stem biomass.
In all but one of our greenhouse trials (with the SPY-containing constructs pATS and pHVS), we harvested the leaves, stems, and roots separately from randomly selected plants; all plants were harvested from the October 2008 trial with pCES:GA20 and pEXP:GA20. The number of ramets per insertion event harvested varied from 4 to 20 depending on the trial (see Table S2). We oven-dried all harvested shoots (leaves and stems) and roots and measured dry weight. We calculated root fraction (root biomass divided by total biomass) for assessing whether the constructs caused cumulative changes in root vs. shoot allocation of growth.
Sequential harvest study
Han et al. (2010) reported that PtGA20-imparted growth modifications differed between early and late stages of tree development. To test whether there was differential biomass allocation during development in GA-modified trees, we sequentially harvested, dried, and measured the roots, leaves, and stems from a subset of plants in the August 2009 greenhouse trial. This study involved six pRGL:GA20-transformed events and six pGA2:GA20-transformed events (5–6 ramets per event), along with both wild-type and empty-vector controls. The organs were harvested every 8–13 days (on May 21, 2010, June 3, 2010, June 14, 2010, and June 22, 2010, respectively) and oven-dried then weighed. Stem, leaf, and root fractions of total biomass were used for statistical analyses as discussed below. Greenhouse conditions were essentially the same as summarized above.
Gibberellin measurements
To assess if there was evidence for changes in GA levels from our constructs, we selected the pGA20 and pPHOR transformants from our July 2009 greenhouse trial for analysis. We randomly collected samples (two young, 4–5-cm leaves per tree) from six ramets per cisgenic event and eight wild-type controls. We then immediately froze them in liquid nitrogen and subsequently freeze dried the samples. We then pooled the samples (creating two pools per type) and then sent them for analysis at the University of Lethbridge, Canada. In short, the leaves were ground with a Retsch MM200 ball mill and then ∼0.3 g subsamples were extracted in 80 % MeOH with internal standards of [2H2]- GA53, GA44, GA19, GA20, GA29, GA1, GA8, GA4 and GA34 (Prof. L. Mander, Australian National University) (Fig. 1). The GAs were purified and measured essentially as described in Busov et al. (2006) except that the methylated samples were trimethylsilylated and analyzed by GC-SIM after NH2 SPE without intervening HPLC. This permitted simultaneous analysis of the 13-hydroxylatedGAs and the final non-13-hydroxylated GAs of poplar (Fig. 1; Pearce et al. 2002).
Field trial design and measurements
We conducted four separate field trials with six different constructs (Table S3): the first with pGA20-transformed plants (planted in summer 2008), the second with pATS- and pHVS-transformed plants (planted in summer 2008), the third with pPHOR-transformed plants (planted in spring 2009), and the fourth with pRGL:GA20- and pGA2:GA20-transformed plants (planted in spring 2009). While the third and the fourth field trials had a CRD, the first two had a RBD (two blocks), with the trees being planted in pairs (row plots). The field trials had 3 to 35 transformed events per construct, with corresponding wild-type and/or empty-vector controls. We tested a total of 92 insertion events covering 556 trees in the field over two growing seasons. Weeds were lightly managed by mowing between rows every 4 to 6 weeks during the growing season. The sites were irrigated during the growing season using overhead sprinkler systems. We recorded tree diameters at 4 and 12 in. from the ground (for pGA20, pATS, and pHVS tested 2008 trials) or height (for pPHOR, pRGL:GA20, and pGA2:GA20 tested 2009 trials), during the dormant season. Diameter squared (the average of diameter2 at 4 in. and diameter2 at 12 in. from ground) or volume index (height × diameter2) was used as a surrogate for total stem biomass.
Statistical analyses
General approach
In analyzing greenhouse and field trials, we first applied Student’s t tests to examine whether the two types of controls (i.e., empty-vector control and wild-type control) were significantly different from each other. If not, we pooled them together to enlarge population size and thus increase the precision of estimated means and statistical tests in subsequent analyses. If there was a significant difference between the two control groups, we used the empty-vector control for comparison to the GA constructs. For the September 2007 greenhouse trial (including pATS and pHVS) and the subsequent field trial in 2008, we only included wild-type plants, therefore used them as controls for analyzing volume index and diameter squared. For the July 2008 greenhouse trial (including pRGL:GA20 and pGA2:GA20), we used wild-type controls when analyzing root fraction as we discovered that the empty-vector control plants had been distributed non-randomly in the experiment.
One-way or two-way ANOVA was used to test if volume index, diameter squared, or root fraction was different in the tested constructs vs. controls. In analyzing greenhouse and field data, we considered events as random effects. For the trials with a CRD, we performed a one-way ANOVA using either volume index, diameter squared, or root fraction as the response variable. For the trials with a RBD, we did a two-way ANOVA with blocking as a fixed effect. If both blocking and the interaction term were non-significant, we ignored blocks and analyzed data as a CRD. For each of the chosen models, we examined plotted residuals vs. fitted values to insure compliance with model assumptions (an example figure is shown in Fig. S3). For several trials, the data were log-transformed to meet equal variance assumptions (this was done for the October 2008 greenhouse trial with pEXP:GA20 and pCES:GA20 and field trials with pATS, pHVS, pRGL:GA20, and pGA2:GA20). The type of controls, ANOVA and parameters, and transformations of data used for analyzing greenhouse trials and field trials are summarized in Tables S4 and S5, respectively. We performed Spearman rank correlation analysis on event means of volume index to examine the similarity of event performance between repeat greenhouse tests, and between greenhouse and field tests. All statistical analyses were carried out using R (version 3.1.2, 2014).
Sequential harvest study
In analyzing data from the sequential harvest study, we pooled the two types of controls together as they were not significantly different from each other based on Student’s t tests. We first did a one-way ANOVA by harvest with shoot fraction as the response variable. Then, using the data from the last three harvests (to avoid potential effects of transplant shock on the first harvest), we modeled the relationship of shoot, root, leaf, or stem biomass to total biomass using weighted regression (Zuur et al. 2009) to deal with observed heteroscedasticity. In this analysis, we included event as a random effect. We excluded five outlier events (one event from the pooled control group, three from population transformed with pGA2:GA20, and one from population transformed with pRGL:GA20) from the regression analysis due to either unusually low total biomass or high root biomass (Fig. S4).
GA levels
We analyzed changes of GA1, GA8, and GA20 concentrations in pGA20- and pPHOR-transformed plants vs. wild-type control using MANOVA and individual ANOVAs. Data were log transformed to meet the equal variance assumption. For this analysis, we were interested in the changes of bioactive GA1 and its direct biosynthetic precursor, GA20, and its 2-hydroxylated catabolite, GA8. We examined correlation (using Pearson r correlation) between volume index and GAs that showed significantly higher levels in cisgenic plants, and tested the ratios of GAs to their direct biosynthetic precursor using ANOVA and Tukey’s HSD. In analyzing GA8/GA1 ratios, a weighted regression model was used to allow for heterogenous variances among groups. An outlier from pGA20 events was dropped during analysis of GA20/GA19 because of its unusually high GA level.
Results
Promoter activity
To verify activity and conduct a qualitative analysis of tissue specificity of expression of the promoters used to drive the PtGA20ox2-2 gene, we collected tissues from plants transformed with each of the promoters driving a GUS reporter gene and performed histochemical analysis (Table S6 and Fig. S5). The GA20 and CES promoters showed strongest expression in fully elongated or elongating internodes, with average scores of 1.3 and 2.3, respectively. The EXP promoter showed its strongest expression in mature root sections (average score of 1.2). The GA2 promoter showed highest activity in shoot tips and young leaves, with an average score of 1.8 in these tissues. All promoters produced highly consistent expression patterns (Fig. S6). For example, all of the pGA20:GUS- and pCES:GUS-transformed events (16 each) showed expression in fully elongated or elongating internodes, 82 % of pEXP:GUS-transformed events showed expression in mature root sections, and 80 % of pGA2:GUS-transformed events showed expression predominantly in shoot tips and young leaves.
For promoters GA20, CES, and EXP, GUS expression was consistent with expectation based on microarray data for their associated native genes (Table S7). The GUS-assay-based specificity index (specificity indexG) showed that the GA20 promoter had the highest specificity (1.9), followed by CES (1.3), and EXP (1.1). The GA2 promoter, however, showed higher specificity in leaves (specificity indexG of 1.3) rather than in stems as had been predicted (specificity indexR of 1.7).
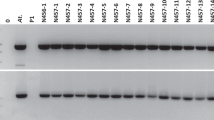
Because we were unable to analyze the expression pattern of promoter RGL with GUS assay due to a construct error (see “Materials and methods”), we verified its activity in pRGL:GA20-transformed plants by study of its expression when fused to the PtGA20ox2-2 protein coding gene (Fig. S7). Semi-quantitative RT-PCR revealed bands in leaf samples from all of the six tested pRGL:GA20-transformed events, but no bands in leaf samples from the transgenic control plants transformed with the pBART empty vector (Fig. S7c). qRT-PCR analysis confirmed the expression of the intragene driven by the RGL promoter in leaf samples, with up to 12-fold greater expression in pRGL:GA20-transformed events compared to those transformed with pBART (the empty vector control) (Fig. S7d). We detected very little expression in shoot tips in four of the tested pRGL:GA20-transformed events; however, the other two events had an average of 5-fold higher expression of the intragene compared with that in the control (Fig. S7e).
Genetic modifications change biomass growth rate and allocation under greenhouse conditions
To examine the performance of all transformed poplars under controlled conditions, we performed six greenhouse trials from 2007 to 2009 (Table S2). The entire population of transformed plants appeared normal and healthy in the greenhouse during all trials, with no physiological or morphological abnormalities when compared with the wild-type control (Fig. 3a, b).
Recombinant DNA (rDNA) modified poplars in greenhouse and field trials. a pGA20- and pPHOR-cisgenic plants during their second week of growth in greenhouse. b pRGL:GA20- and pGA2:GA20-intragenic plants during their fifth week of growth in greenhouse. c pGA20-cisgenic poplars in field. Images were taken on July 6, 2009, Aug 14, 2008, and Aug 19, 2010, respectively
When considering all transformed events, seven out of the eight tested constructs conferred higher volume index (white bars in Fig. 4); the improvement varied from 18 to 146 %. pGA20, pPHOR (July 2009), pRGL:GA20 (July 2008), and pGA2:GA20 (July 2008) caused a statistically significant improvement in volume index (Table 2), in some cases strikingly so (e.g., pRGL:GA20 tested in July 2008; Fig. 5). When considering only the top 25 % of the transformed events, all of the eight tested constructs induced the growth improvement, which varied between 36 and 227 %, with plants transformed with the pGA20 and pRGL:GA20 (July 2008) constructs showing more than 100 % growth improvement (black bars in Fig. 4). However, the results among different greenhouse trials were variable. For example, the high growth improvement (above 50 %) conferred by the pRGL:GA20 and the pGA2:GA20 constructs in the trial done in July 2008 was not observed in the later trial done in August 2009, where plants transformed with the two constructs showed a slight decrease in volume index, while in the case of pPHOR, events in the July 2009 trial showed greater growth improvement compared with those in the June 2008 trial (Fig. 4 and Table 2). The Spearman rank correlation tests, measuring the stability of event performance between repeat greenhouse tests, was significant for the pGA2:GA20 (p = 0.016) but non-significant for pPHOR or pRGL:GA20 transformants (Table S8), suggesting inconsistent growth performance of events within these latter two constructs.
Relative change in volume index of eight constructs tested in the greenhouse. Details on quantitative comparisons made are in Table S4. Years (08 = 2008, 09 = 2009) after constructs indicate plants transformed these constructs were tested twice. Bars denote standard errors for percentage differences of events from control means. The asterisks indicate statistically significant differences (***p < 0.001; *p < 0.05)
We also calculated root fraction to determine whether the different rDNA modifications caused changes in biomass allocation to roots vs. shoots. Four constructs caused apparent reductions in root fraction compared with controls (Fig. 6). The reduced root fractions in pRGL:GA20-, pGA2:GA20-, pGA20-, and pCES:GA20-transformed plants varied from −16 to −7 %; however, only the reduction caused by pGA20 was statistically significant (Table 2; p = 0.012). pPHOR-transformed plants showed a 5 % increase in root fraction in the June 2008 trial but had an almost identical root fraction as controls in the July 2009 trial (Fig. 6). During the October 2008 trial, however, pEXP:GA20 imparted a highly significant 19 % improvement (Fig. 6 and Table 2; p = 0.003) in root fraction.
Root fraction of rDNA modified plants tested in the greenhouse. Years (08 = 2008, 09 = 2009) after constructs indicate plants transformed these constructs were tested twice. Bars denote standard errors for percentage differences of events from control means. The asterisks indicate statistically significant differences (**p < 0.01; *p < 0.05)
Intragenes affect dynamics of biomass allocation
To better understand the variation and dynamics in biomass allocation, we separately harvested leaf, stem, and root samples from the pGA2:GA20- and pRGL:GA20-transformed plants at four different time periods (called harvests) during the August 2009 trial. These constructs were chosen because they showed higher volume indices but lower root fractions in greenhouse studies conducted in 2008 (see above).
When different organs’ biomasses were analyzed separately, pRGL:GA20-transformed plants had a significantly higher shoot fraction than pooled controls (i.e., combined wild-type and empty-vector controls, discussed under statistical analyses) in the third harvest (p = 0.014; Fig. 7 and Table 3). The pGA2:GA20-transformed plants did not significantly differ from the pooled controls in any of the four harvests. We also compared leaf biomass and stem biomass fractions for each harvest between the two intragenic populations and pooled controls; neither pRGL:GA20- nor pGA2:GA20-transformed plants showed significant differences from the controls, however, plants transformed with pRGL:GA20 differed from those transformed with pGA2:GA20 (p = 0.015, leaf fraction from the second harvest, and p = 0.020, stem fraction from the fourth harvest; Table S9).
To analyze biomass distribution trends over harvests, we performed weighted regression to model the relationship between shoot, stem, and root biomass vs. total biomass growth. pRGL:GA20-transformed plants allocated significantly more biomass to their shoots compared to controls (p < 0.001; Fig. 8 and Table 4); for every 100 g increase in total biomass, there was an expectation that 69.4 g would be distributed to shoots in the pooled control group but 73.6 g in the pRGL:GA20 group (a difference of 6 %). The distribution of total biomass to shoots in pGA2:GA20-transformed plants did not differ significantly from the pooled controls (p = 0.082; Fig. 8 and Table 4). When leaf and stem biomass vs. total biomass was each analyzed, both pRGL:GA20- and pGA2:GA20-transformed plants allocated significantly more biomass to their leaves than did controls (Fig. S8 and Table S10); however. there were no significant differences in stem fraction among the three groups (Fig. S9 and Table S10).
Regression lines indicating the relationships between shoot biomass (grams dry weight) and total plant biomass growth in all genotypes (left) and then pooled controls (red diamond), pGA2:GA20 (green triangle), and pRGL:GA20 (blue square) transformed plants. The gray shading shows 95 % confidence intervals (statistics shown in Table 4)
We compared residual variances to examine if GA intragenes increased developmental instability (Table 5 and S11). The results indicated that pRGL:GA20, but not pGA2:GA20, conferred more shoot to total biomass fraction growth variance than did controls; however, no significant differences were found for leaf or stem fraction compared to controls for either construct.
Cisgenes affect gibberellin concentrations
We measured the levels of nine different GAs (Fig. 1) in our pGA20- and pPHOR-transformed poplars to test if a sample of the tested cisgenes affected GA at a detectable level. The transformed poplars showed similar trends in relative GA concentrations to that in wild-type controls. For example, the main bioactive GA present in both wild-type and transformed poplars was GA1. It appeared that GA4 was present, but it was not possible to measure it reliably because of low and/or comingled signals. Additionally, in wild-type and transformed plants, the bioactive GA1 (black bars in Fig. 9a) had lower concentrations than its precursor GA20 (white bars in Fig. 9a) and its catabolite GA8 (black bars in Fig. 9b). GA20, the precursor to the bioactive GA1, showed lower concentrations than its catabolite, GA29 (white bars in Fig. 9b).
Gibberellin (GA) concentrations in leaves of pGA20- and pPHOR-transformed plants compared with wild-type (WT) controls. a The concentrations of bioactive GA1 compared to its precursor GA20. b The concentrations of the inactive catabolites, GA29 (from GA20) and GA8 (from GA1). Note the different y-axis scales. Bars denote standard error over replicate determinations from pooled tissue samples (Table 6). The asterisks indicate statistically significant differences (***p < 0.001; **p < 0.01)
When compared with wild-type control, we observed a 1.2- to 2.9-fold increase in all of the measured GAs in the transformed poplars. In the case of C19 GAs (GA20, GA1, GA8, GA29, and GA34), pGA20-cisgenic plants had the highest concentration, followed by pPHOR-cisgenic plants and the controls (Table 6). MANOVA (focusing on GA1 and its direct precursor GA20 and catabolite GA8, discussed in statistical analyses) indicated GA1 and GA20, significantly differed at the genotype level based on Wilks’ lambda (p = 0.005 for GA1, p = 0.013 for GA20, and p = 0.070 for GA8). ANOVA’s of individual GAs showed that pGA20-transformed plants had a significantly higher concentration of both GA20 (p = 0.048) and GA1 (p = 0.030) than control plants. However, neither of GA20 nor GA1 concentration was significantly correlated with volume index in GA20 transformed plants (correlation coefficient = 0.78 and p = 0.06 for GA20; correlation coefficient = 0.35 and p = 0.82 for GA1). The pPHOR-transformed plants did not significantly differ from controls in any of the three GAs.
The negative and positive feedback regulation known for GA20ox, GA3ox, and GA2ox help to establish GA homeostasis (Fig. 1), and this could be a major factor that prevents tested genes from substantially disturbing GA metabolism. We therefore examined the effect of pGA20 and pPHOR on the 20-, 3-, and 2-oxidation processes with a focus on the conversions (i.e., ratios) between each GA and its direct biosynthetic precursor. Compared with control, pGA20- and pPHOR-transformed plants showed same trends in changes of the ratios examined (Fig. S10). Two exceptions were GA44/GA53 and GA20/GA19, where pGA20 conferred higher ratios while pPHOR conferred slightly lower ratios to control. The ratios of GA8/GA1 and GA29/GA20, regulated by the GA2ox, were significantly lower in pGA20- and/or pPHOR-transformed plants than those in controls (p = 0.0163 for GA8/GA1 in pGA20; p < 0.001 for GA29/GA20 in pGA20; p = 0.005 for GA29/GA20 in pPHOR). No significant changes were found in GA conversions regulated by GA20ox or GA3ox.
Field studies
We performed four field trials (two in the summer of 2008 and two in the spring of 2009) to examine the field growth of poplars modified with six of the tested constructs (Table S3). As was the case with our greenhouse trials, all of the trees growing in the field looked normal and healthy (Fig. 3c).
Trees transformed with pRGL:GA20, pGA2:GA20, and pATS showed a noticeably higher volume index (or diameter squared) when compared with controls (white bars in Fig. 10); the improvement conferred by pRGL:GA20 was statistically significant (p = 0.04; Table 7). When considering only the top 25 % of the transformed events (as might be chosen for possible application in a breeding program), the growth improvement varied between 13 and 89 %, with plants transformed with the pRGL:GA20 and pATS constructs showing more than 50 % growth improvement (black bars in Fig. 10). When comparing results from the greenhouse and the field, we obtained generally similar trends, with notable exceptions being pGA20-, pPHOR-, and pHVS-transformed plants. pGA20-transformed plants showed a 146 % volume index improvement (significant) in the greenhouse but had almost identical volume index as the controls under field conditions. pPHOR-transformed plants that had a 26 % volume index improvement (non-significant) in the greenhouse had a 22 % reduction in average volume index in the field. A similar trend was observed for pHVS-transformed plants. Growth in the field did not correlate with growth in the greenhouse. Spearman rank correlation analysis of volume index between events grown in both greenhouse and field tests was significant for only pRGL:GA20 (tested in 2009 field and July 2008 GH trials; p = 0.042), but non-significant for other constructs (Table S12).
Discussion
We report on a number of overlapping studies, conducted over 6 years, that investigated the extent to which a variety of genetic perturbations of GA metabolism or signaling might be useful tools for modifying tree growth rate and allocation of biomass. We tested eight promoter-coding region combinations of GA pathway components, including two cisgenes (pGA20 and pPHOR), four intragenes (pRGL:GA20, pGA2:GA20, pEXP:GA20, and pCES:GA20), and two transgenes (pATS and pHVS), for their ability to alter GA homeostasis and growth in poplar. Our results suggest that modified GA levels in poplar can indeed lead to changes in either shoot biomass or its allocation among organs, while at the same time produce plants with relatively normal physiology and adaptation (e.g., lacking severe pleiotropic phenotypes).
Our characterization of GA concentrations suggest that GAs from the 13-hydroxylation pathway are the dominant forms in our wild-type and cisgenic plants, and GA1, rather than GA4, is the major bioactive gibberellin. These findings are consistent with our earlier characterization of GA concentrations in wild-type and gai- and rgl1-transgenic poplars, where the level of GA1 was much higher than that of GA4 (Busov et al. 2006). However, Eriksson et al. (2000) and Israelsson et al. (2004) found higher levels of GA4 than GA1 in the leaves of both wild-type and transgenic poplars. These differences may be due to the use of difference poplar clones (Populus tremula L. × P. tremuloides Michx. clone T89 in their studies, vs. 717-1B4 P. tremula × P. alba in our studies), or growth conditions. The source of the GA20ox gene may also contribute to the differences; Eriksson et al. (2000) used the GA20ox gene derived from Arabidopsis (AtGA20ox1) to induce overexpression, whereas we used a native poplar gene (PtGA20ox7).
Although the effects of feedback regulation of 2-ODDs could have prevented plants from producing excessive amounts of bioactive GAs, we were still able to observe increased levels of each of the GAs we measured, as well as disturbed GA homeostasis in the pGA20- and pPHOR-cisgenic poplars. The concentrations of GA20 and GA1 in pGA20-transfromed plants were significantly higher than those in controls; this alone could be an explanation for the decreased in the ratios of GA29/GA20 and GA8/GA1 in pGA20-transformed plants. The decrease in the two ratios could also be due to a slower turnover from the GA2ox substrates to the products, since similar changes were also observed in pPHOR-transformed plants, where the levels of GA2ox substrates showed no difference from the control. In contrast to our observations, however, Eriksson et al. (2000) reported decreased levels of GA20ox substrates (GA53 and GA19) and sharply increased GA8/GA1 ratios in their AtGA20ox1-transgenic poplar clones. A reason could be the CaMV 35S promoter used in their study. It possibly led to very strong expression of the transgene, and therefore, severely disturbed the GA pathway and its feedback regulation.
Modified expression of genes encoding GA biosynthetic enzyme (GA20ox) and signaling regulators (PHOR and SPY) did not cause any notable morphological changes in either greenhouse or field conditions, but did lead to strong growth improvements in some experiments. Two poplar PHOR1 orthologues, PtPHOR1-1 and PtPHOR1-2, have been identified previously (Zawaski et al. 2012). Unlike PtPHOR1-1 which is short-day inducible and preferably expressed in roots, the PtPHOR1-2 gene tested in this study has been suggested to be expressed independently of short-day treatment and uniformly in leaf, stem, and root. Similar to our observation, significant growth improvement in shoots and roots in PtPHOR1-2 overexpressing poplars has also been observed by Zawaski et al. (2012), where the gene was driven by the CaMV 35S promoter and constitutively expressed.
Under greenhouse condition, the observed effects from the genes largely agreed with our expectations. For example, the EXP promoter (PtEXPB1) was chosen because it is mainly expressed in roots (microarray data produced by Rodgers-Melnick et al. 2012). The significantly increased root fraction in pEXP:GA20-transformed plants suggests an increased expression of GA20ox in roots. PtEXPB1 belongs to the β-expansin (EXPB) gene family, whose expression pattern remains unexplored in poplar. Like other expansins, EXPBs are cell wall-loosening proteins. Studies in several plant species have suggested tissue-specific expression patterns and diverse biological functions of EXPBs. In maize, where EXPBs were firstly identified as group-1 grass pollen allergens, some EXPBs (e.g., Zeam1) are known to play a role in pollen tube penetration (Cosgrove et al. 1997), while others (e.g., ZmEXPB2) appear to take part in root elongation (Kam et al. 2005). Root-specific/preferred EXPBs have also been isolated from barley (HvEXPB1), wheat (TaEXPB23), rice (OsEXPB5), Arabidopsis (AtEXPB1), and soybean (GmEXPB1, GmEXPB2, and GmEXPB6) (Kwasniewski and Szarejko 2006; Gao et al. 2008; Kwon et al. 2008; Won et al. 2010; Li et al. 2011, 2013). The root-specific activity of HvEXPB1 from barley and OsEXPB5 from rice has been shown to be conferred by root hair specific cis-elements (RHEs) (Won et al. 2010).
We were surprised by the growth advantage conferred by both the SPY constructs. As mentioned earlier, SPY is a negative regulator of GA signaling; the expectation from using these constructs was that we would produce dwarf plants. The results, however, were the opposite: SPY genes from both Arabidopsis (AtSPY) and barley (HvSPY) conferred growth improvements in the transgenic poplars. Interestingly, both SPY orthologs have caused growth retardation as well as acceleration in other plant species. Overexpression of AtSPY inhibited shoot elongation in Petunia (Izhaki et al. 2001), but promoted hypocotyl, rosette, and stem growth in Arabidopsis (Swain et al. 2001). The HvSPY adopted in our study, although with partial deletion in the eighth and ninth tetratricopeptide repeats (TPRs), retains SPY function (Filardo et al. 2009). By overexpressing the modified HvSPY, Filardo et al. (2009) observed reduced height but increased rosette size in spy-4 Arabidopsis, which usually has a long stem but small rosette size; however, they did not see clear morphological alterations in wild type Arabidopsis. The variations in SPY-induced phenotypic changes are believed due to its additional roles in cytokinin and redox signaling, and consequent effects on plant root development, brassinosteroid synthesis, circadian rhythm and photoperiodic flowering, and abiotic stress response (Tseng et al. 2004; Swain et al. 2001; Shimada et al. 2006; Filardo et al. 2009; Qin et al. 2011; Steiner et al. 2012; Cui et al. 2014).
Studies of biomass allocation showed that the increase in biomass growth we observed was associated with preferential allocation to shoots. pGA2:GA20- and pRGL:GA20-transformed plants showed the highest volume indices but produced lower root fractions. pGA2:GA20-transformed plants showed increased leaf mass fraction, and decreased stem mass and root mass fractions. This was consistent with previous results showing that although GAs are positive regulator of root growth, they significantly repress root development (Eriksson et al. 2000; Busov et al. 2006; Gou et al. 2010, 2011; Jeon et al. 2015). The increase in leaf mass fraction in our pGA2:GA20-intragenic plants was comparable to the slight increase shown by Eriksson et al. (2000). Likewise, pRGL:GA20-transformed plants showed increased leaf and shoot mass fractions, and decreased root mass fraction. This suggests that pGA2:GA20- and pRGL:GA20-tranformed plants allocated greater biomass to shoots under greenhouse conditions, where plants had a stable temperature regime, no wind or insolation stress, and did not suffer water or nutrient deficiencies. The modified biomass allocation in plants transformed with pRGL:GA20 was further confirmed in our sequential harvest study, where we found that the pRGL:GA20-transformed plants grew faster and allocated significantly greater biomass to their leaves. Overall, the changes in biomass allocation we observed—with preferential root mass or shoot mass obtained with different constructs—if expressed in the field could rise to large cumulative differences in relative organ size in trees, with potential impacts on tree growth rate, stability, wind-firmness, drought tolerance, and capacity for bioremediation (Klocko et al. 2013). These possible effects require evaluation under experimental manipulations, and in long-term field trials.
To our surprise, retesting many of the same events in subsequent greenhouse trials failed to show consistent levels of growth enhancement. Initial testing of pPHOR-, pRGL:GA20-, and pGA2:GA20-transformed plants were started in June or July in 2008 and lasted for 2 months. Subsets of events, representing high, median, and low growth rates, were tested again for 1 month, starting in July or August in 2009. Although we tried to keep greenhouse conditions similar among the two trials, including water and fertilizer regimes, greenhouse conditions still vary widely, particularly the levels of light and high and low temperatures. Although performances of pGA2:GA20-transformed events were correlated in the two tests, improved performance was not seen in the second test, and selective testing of specific pGA2:GA20-transformed events did not replicate the ranking in growth improvement seen in the original pGA2:GA20-intragenic population. The pGA2:GA20 construct might have conferred a benefit for early growth that dissipated during later stages of development, similar to that reported by Han et al. (2010). It is also possible that developmental changes that occurred as a result of many months of in vitro growth, or responses to the stress during re-acclimation and re-propagation in the greenhouse, resulted in the reduction of GA expression or the loss of responsiveness to GA overproduction.
The inconsistent growth performance seen between replicated greenhouse trials of the same events, and among greenhouse and field tests of the same events, could also have resulted from instability of transgene expression, which has been observed in poplars under in vitro, greenhouse, and field conditions (Kumar and Fladung 2001). It can result from gene loss, transcriptional gene silencing (TGS), or post-transcriptional gene silencing (PTGS) (Kooter et al. 1999; Fagard and Vaucheret 2000). Although high stability of transgene expression has been reported in a number of studies of transgenic poplars (reviewed by Brunner et al. 2007; Li et al. 2009), most studies have been of exogenous genes and promoters which do not resemble those present in the poplar genome. It is well known that extra copies of promoters and genes can trigger or increase methylation and homology-dependent silencing (Matzke et al. 2000). As very similar versions of most of the promoters and genes tested in our study were also present within the poplar genome, they may have been subject to TGS and PTGS. It will be of interest to further examine this, and to compare stability of trait expression when exogenous promoters and coding regions are used vs. native (cisgenic or intragenic) ones as tested here.
Genotype by environment interaction, as well as low statistical precision in these exploratory research trials, could also have been a major reason for the poor correlation among greenhouse and field performance. In the greenhouse, plants grow in a controlled environment where all environmental factors such as light, humidity, and temperature are controlled and mild. Plants are also fertilized and watered regularly, and do not have to compete with weeds for resources such as minerals and water. In the field, however, plants are subject to much more stressful and variable environmental conditions. Abiotic stresses, including cold, salt, and osmotic stresses, have been shown to affect the transcription level of RGL3 and several GA2ox genes in Arabidopsis (reviewed by Colebrook et al. 2014). It is very likely that the promoters (e.g., RGL and GA2) employed in our study also show environmental instability, which would affect the transcription of GA20ox and consequently GA levels. The exposure to more extreme (low and high) temperatures could have led to differential reduction in bioactive GAs, similar to that observed in Arabidopsis and rice (Oryza sativa) (Toh et al. 2008; Colebrook et al. 2014; Sakata et al. 2014). GA signal transduction pathways strongly interact with phytohormone-regulated stress response pathways (reviewed by Weiss and Ori 2007; Colebrook et al. 2014), including antagonistically with the abscisic acid (ABA) pathway. It is well known that ABA is a central hormone that helps to organizes plant responses to environmental stresses (Hofmann 2008; Finkelstein 2013). The stresses in the field, for example reduced water availability, are likely to have elevated ABA production, in turn inhibiting GA effects. GAs have also been shown to negatively interact with ethylene signal transduction pathways; ethylene also plays a key role in organizing plant responses to various stresses (Hall and Smith 1995; Achard et al. 2003). The antagonistic interaction of ethylene and GA might also lead to suppression of the growth promoting effects of bioactive GAs.
The inconsistent growth enhancement and strong genotype by environment interaction observed in our study emphasizes the need for extensive field testing in applied biotechnology research. However, under current stringent laws and regulations pertaining to genetically modified crops, studies are either restricted to greenhouse testing alone (e.g., Eriksson et al. 2000; Han et al. 2010; Zawaski et al. 2012; Jeon et al. 2015), or confined to small-scale field trials for a minimum duration of time to minimize or avoid any flowering (e.g., 3 years in Jing et al. 2004; only 1 year in Kim et al. 2011). Policy changes are likely to be needed to provide a more conducive environment for field trials of transgenic trees if growth rate and plant architecture-modifying transgenes, especially cisgenes and intragenes whose effects are comparable to that from conventional breeding, are to be used in breeding programs (Strauss et al. 2015).
Finally, our work demonstrates both the benefits and risks of cis/intragenic approaches for disturbing endogenous GA levels and modifying architecture in poplars. Unlike poplars containing genes from sexually incompatible species (like those produced by Eriksson et al. 2000), most of the recombinant DNA-modified poplars tested in our study were similar in DNA sequence to those from conventional breeding, thus may be better accepted by the public (Holme et al. 2013). Additionally, cis/intragenic crops have been proposed to be excluded from GMO regulatory frameworks or regulated at a lower level of stringency, possibly avoiding much of the costs and stigma of transgenic GMOs (Schouten et al. 2006; Strauss et al. 2015). However, the instability of trait expression we observed suggests that cis/intragenics, at least for complex traits like hormone-based growth modification, might possibly hold risks of trait instability that are larger than those for transgenics. Additional research, particularly field studies, are needed to understand this possible dilemma.
References
Achard P, Genschik P (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 60(4):1085–1092
Achard P, Vriezen WH, Straeten DVD, Harberd NP (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15(12):2816–2825
Amador V, Monte E, García-Martínez JL, Prat S (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to drosophila armadillo. Cell 106(3):343–354
Bhattacharya A, Kourmpetli S, Davey MR (2009) Practical applications of manipulating plant architecture by regulating gibberellin metabolism. J Plant Growth Regul 29(2):249–256
Brunner A, Li J, DiFazio SP, Shevchenko O, Mohamed R, Montgomery B, Mohamed R, Wei H, Ma C, Elias AA, VanWormer K, Strauss SH (2007) Genetic containment of forest plantations. Tree Genet Genomes 3:75–100
Busov VB, Meilan R, Pearce DW, Rood SB, Ma C, Tschaplinski TJ, Strauss SH (2006) Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 224(2):288–299
Busov VB, Brunner AM, Strauss SH (2008) Genes for control of plant stature and form. New Phytol 177(3):589–607
Chabaud M, De Carvalho-Niebel F, Barker DG (2003) Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell Rep 22(1):46–51
Claeys H, De Bodt S, Inzé D (2014) Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci 19(4):231–239
Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217(1):67–75
Cosgrove DJ, Bedinger P, Durachko DM (1997) Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci 94(12):6559–6564
Cui H, Kong D, Wei P, Hao Y, Torii KU, Lee JS, Li J (2014) SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox-mediated cortex proliferation in the Arabidopsis root. Mol Plant 7(12):1727–1739
Davière JM, Achard P (2013) Gibberellin signaling in plants. Development 140(6):1147–1151
Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18(7):784–788
Fagard M, Vaucheret H (2000) (TRANS)GENE SILENCING IN PLANTS: how many mechanisms? Annu Rev Plant Physiol Plant Mol Biol 51(1):167–194
Filardo F, Robertson M, Singh DP, Parish RW, Swain SM (2009) Functional analysis of HvSPY, a negative regulator of GA response, in barley aleurone cells and Arabidopsis. Planta 229(3):523–537
Filichkin SA, Meilan R, Busov VB, Ma C, Brunner AM, Strauss SH (2006) Alcohol-inducible gene expression in transgenic Populus. Plant Cell Rep 25(7):660–667
Finkelstein R (2013) Abscisic acid synthesis and response. The Arabidopsis Book, e0016
Gallego-Giraldo L, Ubeda-Tomás S, Gisbert C, García-Martínez JL, Moritz T, López-Díaz I (2008) Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant Cell Physiol 49(5):679–690
Gao Q, Zhao M, Li F, Guo Q, Xing S, Wang W (2008) Expansins and coleoptile elongation in wheat. Protoplasma 233(1–2):73–81
García-Hurtado N, Carrera E, Ruiz-Rivero O, López-Gresa MP, Hedden P, Gong F, García-Martínez JL (2012) The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J Exp Bot 63(16):5803–5813
Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20(6):1203–1207
Gou J, Strauss SH, Tsai CJ, Fang K, Chen Y, Jiang X, Busov VB (2010) Gibberellins regulate lateral root formation in populus through interactions with auxin and other hormones. Plant Cell 22(3):623–639
Gou J, Ma C, Kadmiel M, Gai Y, Strauss SH, Jiang X, Busov VB (2011) Tissue-specific expression of Populus C19 GA 2-oxidases differentially regulate above- and below-ground biomass growth through control of bioactive GA concentrations. New Phytol 192(3):626–639
Hall MA, Smith AR (1995) Ethylene and the responses of plants to stress. Bulg J Plant Physiol 21(2–3):71–79
Han KM, Dharmawardhana P, Arias RS, Ma C, Busov VB, Strauss SH (2010) Gibberellin-associated cisgenes modify growth, stature and wood properties in Populus. Plant Biotechnol J 9(2):162–178
Hedden P, Phillips AL (2000a) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5(12):523–530
Hedden P, Phillips AL (2000b) Manipulation of hormone biosynthetic genes in transgenic plants. Curr Opin Biotechnol 11(2):130–137
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444(1):11–25
Hofmann N (2008) Abscisic acid–mediated suppression of systemic acquired resistance signaling. Plant Cell 20(6):1425
Holme IB, Wendt T, Holm PB (2013) Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol J 11(4):395–407
Israelsson M, Mellerowicz E, Chono M, Gullberg J, Moritz T (2004) Cloning and overproduction of gibberellin 3-oxidase in hybrid aspen trees. Effects on gibberellin homeostasis and development. Plant Physiol 135(1):221–223
Izhaki A, Swain SM, Tseng T, Borochov A, Olszewski NE, Weiss D (2001) The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J 28(2):181–190
Jeon HW, Cho JS, Park EJ, Han KH, Choi YI, Ko JH (2015) Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol J. doi: 10.1111/pbi.12484
Jing ZP, Gallardo F, Pascual MB, Sampalo R, Romero J, De Navarra AT, Cánovas FM (2004) Improved growth in a field trial of transgenic hybrid poplar overexpressing glutamine synthetase. New Phytol 164(1):137–145
Kam MJ, Yun HS, Kaufman PB, Chang SC, Kim SK (2005) Two expansins, EXP1 and EXPB2, are correlated with the growth and development of maize roots. J Plant Biol 48(3):304–310
Kim YH, Kim MD, Choi YI, Park SC, Yun DJ, Noh EW, Lee HS, Kwak SS (2011) Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotechnol J 9(3):334–347
Klocko AL, Stanton B, Oosten C, Strauss SH (2013). Green revolution plantations: could short trees be a big thing?. ISB News Report, May 2013: 7–9
Kooter JM, Matzke MA, Meyer P (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci 4(9):340–347
Kumar S, Fladung M (2001) Gene stability in transgenic aspen (Populus). II. Molecular characterization of variable expression of transgene in wild and hybrid aspen. Planta 213(5):731–740
Kwasniewski M, Szarejko I (2006) Molecular cloning and characterization of β-expansin gene related to root hair formation in barley. Plant Physiol 141(3):1149–1158
Kwon YR, Lee HJ, Kim KH, Hong SW, Lee SJ, Lee H (2008) Ectopic expression of Expansin3 or Expansinβ1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol Lett 30(7):1281–1288
Li J, Brunner AM, Meilan R, Strauss SH (2009) Stability of transgenes in trees: expression of two reporter genes in poplar over three field seasons. Tree Physiol 29(2):299–312
Li F, Xing S, Guo Q, Zhao M, Zhang J, Gao Q, Wang G, Wang W (2011) Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J Plant Physiol 168(9):960–966
Li X, Zhao J, Walk TC, Liao H (2013) Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Appl Microbiol Biotechnol 98(6):2805–2817
Matzke MA, Mette MF, Matzke AJM (2000) Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol 43(2–3):401–415
Mittal A, Decker SR (2013) Special issue: application of biotechnology for biofuels: transforming biomass to biofuels. 3. Biotech 3(5):341–343
Olszewski N, Sun T, Gubler F (2002) Gibberellin signaling biosynthesis, catabolism, and response pathways. Plant Cell Online 14(suppl 1):S61–S80
Otani M, Meguro S, Gondaira H, Hayashi M, Saito M, Han DS, Nakano M, Mori S, Li T, Niki T, Nishijima T, Koshioka M (2014) Overexpression of the gibberellin 20-oxidase or gibberellin 3-oxidase gene from Torenia fournieri affecting plant morphology in transgenic Tricyrtis sp. Int Soc Hortic Sci - ISHS 23–30
Pearce DW, Hutt OE, Rood SB, Mander LN (2002) Gibberellins in shoots and developing capsules of Populus species. Phytochemistry 59(6):679–687
Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K (2011) SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol 157(4):1900–1913
Qin X, Liu JH, Zhao WS, Chen XJ, Guo ZJ, Peng YL (2013) Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol Plant-Microbe Interact: MPMI 26(2):227–239
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ Jr, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T (2006) The path forward for biofuels and biomaterials. Science 311(5760):484–489
Rodgers-Melnick E, Mane SP, Dharmawardhana P, Slavov GT, Crasta OR, Strauss SH, Brunner AM, DiFazio SP (2012) Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res 22(1):95–105
Rood S, Hedden P (1994) Convergent pathways of gibberellin A1 biosynthesis in Brassica. Plant Growth Regul 15(3):241–246
Sakata T, Oda S, Tsunaga Y, Shomura H, Kawagishi-Kobayashi M, Aya K, Saeki K, Endo T, Nagano K, Kojima M, Sakakibara H, Watanabe M, Matsuoka M, Higashitani A (2014) Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol 164(4):2011–2019
Schouten HJ, Krens FA, Jacobsen E (2006) Cisgenic plants are similar to traditionally bred plants: International regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep 7(8):750–753
Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48(3):390–402
Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun T (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143(2):987–1000
Sponsel VM, Hedden P (2010) Gibberellin biosynthesis and inactivation. In: Plant Hormones, Springer Netherlands, pp 63–94
Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24(1):96–108
Strauss SH, Costanza A, Séguin A (2015) Genetically engineered trees: paralysis from good intentions. Science 349(6250):794–795
Swain SM, Tseng T, Olszewski NE (2001) Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol 126(3):1174–1185
Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, Iuchi S, Kobayashi M, Yamaguchi S, Kamiya Y, Nambara E, Kawakami N (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146(3):1368–1385
Tseng TS, Salomé PA, McClung CR, Olszewski NE (2004) SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16(6):1550–1563
Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144(3):1240–1246
Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14(1):87–100
Won SK, Choi SB, Kumari S, Cho M, Lee SH, Cho HT (2010) Root hair-specific EXPANSIN B genes have been selected for graminaceae root hairs. Mol Cells 30(4):369–376
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59(1):225–251
Yamaguchi S, Kamiya Y (2000) Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol 41(3):251–257
Zawaski C, Ma C, Strauss SH, French D, Meilan R, Busov VB (2012) PHOTOPERIOD RESPONSE 1 (PHOR1)-like genes regulate shoot/root growth, starch accumulation, and wood formation in Populus. J Exp Bot 63(15):5623–5634
Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM (2009) Mixed effects models and extensions in ecology with R. Springer New York, New York
Acknowledgments
We thank Oregon State University (OSU) consulting statistician Ariel Muldoon for detailed advice on weighted regression analysis. We thank Dr. Steve Swain (CSIRO Plant Industry, Australia) for providing the two SPY transgene constructs (pATS and pHVS) for testing. We thank industrial members of the TBGRC Research Cooperative at OSU, the US Department of Energy’s Poplar Genome-Based Research for Carbon Sequestration in Terrestrial Ecosystems Program (grant nos. DE-FG02-06ER64185 and DE-FG02-05ER64113), the Consortium for Plant Biotechnology Research, Inc. (GO12026-203A), ArborGen, LLC (04412), USDA CSREES (2003-04345), USDA (2004-35300-14687), and the Natural Sciences and Engineering Research Council (Canada -DG to SRood) for financial support.
Data Archiving Statement
DNA sequences of promoters and coding regions reported in this manuscript will be submitted to the NCBI Genbank. Other data will be made available through the Dryad Digital Repository (http://datadryad.org/depositing).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. M. Dandekar
This article is part of the Topical Collection on GMOs
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 24 kb)
Fig. S1
Tissues used in GUS staining analysis. a A pCES:GUS-transformed plant. Six different types of tissues from above-ground were used in GUS staining analysis. b Three different types of tissues from below-ground were used in GUS staining analysis (DOCX 1029 kb)
Fig. S2
Scoring system used to rate the intensity of GUS staining. a 0 = no expression. b 1 = low expression. c 2 = medium expression. d 3 = high expression (DOCX 78 kb)
Fig. S3
Model validation for the chosen regression model of shoot biomass vs. total biomass. Residuals were plotted vs. fitted values, total biomass, construct, and harvest time, respectively (DOCX 2205 kb)
Fig. S4
Five data points were considered as outliers and excluded from regression analysis. a Data from the 55 individual plants in the control group (26 transgenic trees from 4 pBART-transforments and 29 non-transgenic trees) was plotted; 1 data point (in red circle) was dropped and excluded from further analyses due to its low total biomass. b Data from the 108 pGA2:GA20-intragenic plants was plotted; 3 data points were dropped and excluded from further analyses due to their high root biomass (1 data point; in light blue circles) or low total biomass (2 data points; in red circles). c Data from the 106 pRGL:GA20-intragenic plants was plotted; 1 data point (in red circle) was dropped and excluded from further analyses due to its low total biomass (PDF 479 kb)
Fig. S5
Results from GUS staining to determine tissue specificity of promoter expression. a GUS scores of nine different tissue types from pGA20:, pGA2:, pEXP: and pCES:GUS-transgenic plants. Bars denote standard error over biological replicates. b GUS stained root sections from pEXP:GUS-transgenic plants, scale bar = 5 mm. c GUS stained stem sections from pCES:GUS-transgenic plants, scale bar = 5 mm. (ST = shoot tip, YL = young leaf, ML = mature leaf, Pet = petiole, EI = elongating internode, FEI = fully elongated internode, MRS = mature root section, MR = mature root, RT = root tip) (DOCX 148 kb)
Fig. S7
Verification of RGL promoter activity in pRGL:GA20-transformed plants. a Leaf sample collected from pRGL:GA20-transformed plants for RNA isolation. b. Shoot tips collected from pRGL:GA20-transformed plants for RNA isolation. c. Agarose gel analysis of semi-quantitative RT-PCR product from 6 pRGL:GA20 events (numbered as 3, 16, 20, 30, 31 and 38) and empty-vector control (pBART); H2O was included as no template control and gDNA from pRGL:GA20 event 31 was included as a positive control. The size of bands is 167 bp. d. Expression level of GA20 intragene in leaves detected by qRT-PCR. e. Expression level of GA20 intragene in shoot tips detected by qRT-PCT. Bars denote standard error over six replications (two biological replicates with three pseudoreplicates each) (DOCX 671 kb)
Fig. S8
Regression lines indicating the relationships between leaf biomass (grams dry weight) and total biomass in all genotypes (left) and then pooled controls (red diamond), pGA2:GA20 (green triangle) and pRGL:GA20 (blue square) transformed plants. Both pGA2:GA20- and pRGL:GA20-intragenic plants allocated significantly more biomass to their leaves. The gray shading shows 95% confidence intervals (statistics shown in Table S10) (DOCX 419 kb)
Fig. S9
Regression lines indicating the relationships between leaf biomass (grams dry weight) and total biomass in all genotypes (left) and then pooled controls (red diamond), pGA2:GA20 (green triangle) and pRGL:GA20 (blue square) transformed plants. There were no significant differences in stem fraction among the three groups. The gray shading shows 95% confidence intervals (statistics shown in Table S10) (DOCX 404 kb)
Fig. S10
Ratios of GAs to its direct precursor in pGA20, pPHOR and wild-type (WT) control poplars. GA44/GA53, GA19/GA44 and GA20/GA19 are indicators of the 20-oxidation process; GA1/GA20 is indicator of the 3-oxidation process; GA8/GA1 and GA29/GA20 are indicators of the 2-oxidation process. Bars denote standard error over replicate determinations from pooled tissue samples (Table 6). Comparisons were to WT. (***, p <0.001; **, p <0.01; *, p <0.05) (DOCX 44 kb)
Table S1
Primers used to verify transformation (the first nine pairs) and quantify expression (the last two pairs) (DOCX 20 kb)
Table S2
Summary of trials carried out in greenhouse. Exp. No. = experiment number. CRD = completely randomized design, RBD = randomized block design; WT = wild-type control, EV = empty-vector control (DOCX 22 kb)
Table S4
Summary of statistical analyses performed for greenhouse data. Abbreviations in Table S2. “Pooled” indicates two types of controls were combined. Biomass data was not collected from the Sept 07 trial with pATS and pHVS constructs. Root fraction of pRGL:GA20 and pGA2:GA20 from the Aug 09 trial was analyzed in the sequential harvest study (DOCX 23 kb)
Table S6
GUS scores in tested tissues. Brackets represent standard errors over 5 to 17 biological replicates. ST = shoot tip, YL = yong leaf, ML = mature leaf, Pet = petiole, EI = elongating internode, FEI = fully elongated internode, MRS = mature root section, MR = mature root, RT = root tip. Bold numbers indicate the strongest expression for each promoter (DOCX 23 kb)
Table S8
Spearman rank correlation analysis of volume index to examine the similarity of event performance between repeat greenhouse (GH) tests. Experiment number (Exp. No.) of GH tests in Table S2. Bold numbers indicate statistically significant differences (p <0.05) (DOCX 19 kb)
Table S9
Comparison of the mean leaf fractions and the mean stem fractions among pRGL:GA20-, pGA2:GA20-transformed plants and pooled controls. Tukey’s HSD was performed only if ANOVA suggested statistically significant differences among the three groups. Bold numbers indicate statistically significant differences. Data from the first harvest was exluded from analysis to avoid potential effects of transplant shock on plant growth (DOCX 20 kb)
Table S10
Summary of estimated slopes and 95% confidence intervals (CIs) for regression lines. The asterisks indicate statistically significant differences (*, p <0.05) (DOCX 20 kb)
Table S11
Comparisons of residual variances. Levene’s test of homogeneity of variances suggested no statistically significant differences (p <0.05). Data from the first harvest was exluded from analysis to avoid potential effects of transplant shock on plant growth (DOCX 20 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lu, H., Viswanath, V., Ma, C. et al. Recombinant DNA modification of gibberellin metabolism alters growth rate and biomass allocation in Populus . Tree Genetics & Genomes 11, 127 (2015). https://doi.org/10.1007/s11295-015-0952-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0952-0