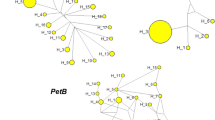
Abstract
Trinidad and Tobago has a long history of producing high-quality cacao (Theobroma cacao L.). Cacao genotypes in Trinidad and Tobago are of a highly distinctive kind, the so-called “Trinitario” cultivar group, widely considered to be of elite quality. The origin of Trinitario cacao is unclear, although it is generally considered to be of hybrid origin. We used massive parallel sequencing to identify polymorphic plastidic single nucleotide polymorphisms (cpSNPs) and polymorphic plastidic simple sequence repeats (cpSSRs) in order to determine the origin of the Trinitario cultivar group by comparing patterns of polymorphism to a reference set of ten completely sequenced chloroplast genomes (nine T. cacao and one outgroup, T. grandiflorum (Willd. ex Spreng.) Schum). Only three cpSNP haplotypes were present in the Trinitario cultivars sampled, each highly distinctive and corresponding to reference genotypes for the Criollo (CRI), Upper Amazon Forastero (UAF) and Lower Amazon Forastero (LAF) varietal groups. These three cpSNP haplotypes likely represent the founding lineages of cacao to Trinidad and Tobago. The cpSSRs were more variable with eight haplotypes, but these clustered into three groups corresponding to the three cpSNP haplotypes. The most common haplotype found in farms of Trinidad and Tobago was LAF, followed by UAF and then CRI. We conclude that the Trinitario cultivar group is of complex hybrid origin and has derived from at least three original introduction events.







Similar content being viewed by others
References
Arroyo-García R, Lefort F, de Andrés MT, Ibáñaez J, Borrego J, Jouve N, Cabello F, Martínez-Zapater JM (2002) Chloroplast microsatellite polymorphisms in Vitis species. Genome 45:1142–1149
Bartley BGD (2005) The genetic diversity of cacao and its utilization. CABI, Wallingford, UK
Bekele FL, Bidaisee GG, Bhola J (2007) A comparative morphological study of two Trinitario groups from the International Cocoa Genebank, Trinidad. Annual Report 2006, Cocoa Research Unit, University of the West Indies, Trinidad and Tobago, pp 34–42
Cheesman EE (1934) The botanical programme of 1933. In Third annual report on cacao research,1933. Government Printing Office, Port-of-Spain, Trinidad, pp 1–2
Cheesman EE (1944) Notes on the nomenclature, classification and possible relationships of cocoa populations. Trop Agric 21:144–159
Coe SD, Coe MD (1996) The true history of chocolate. Thames and Hudson, New York, USA
Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperms. Am J Bot 75:1443–1458
Dieringer D, Schlötterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Engels JMM (1986) The systematic description of cacao clones and its significance for taxonomy and plant breeding. Dissertation, Agricultural University, Wageningen, Netherlands, 125p
Felsenstein J (1973) Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst Zool 22:240–249
Felsenstein J (1989) PHYLIP — phylogeny inference package (Version 3.2). Cladistics 5:164–166
Freeman WE (1969) Some aspects of the cacao breeding programme. Proceedings of Agricultural Society of Trinidad and Tobago, Dec 1968, pp 1–15
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hale ML, Borland AM, Gustafsson MH, Wolff K (2004) Causes of size homoplasy among chloroplast microsatellites in closely related Clusia species. J Mol Evol 58:182–190
Huang CY, Grünheit N, Ahmadinejad N, Timmis JN, Martin W (2005) Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol 138:1723–33
Irish BI, Goenaga R, Zhang D, Schnell R, Brown S, Motamayor JC (2010) Microsatellite fingerprinting of the USDA-ARS tropical agriculture research station cacao (Theobroma cacao L.) germplasm collection. Crop Sci 50:656–667
Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H (2011) Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol 28:835–847
Johnson ES, Bekele FB, Brown SJ, Song Q, Zhang D, Meinhardt LW, Schnell RJ (2009) Population structure and genetic diversity of the Trinitario cacao (Theobroma cacao L.) from Trinidad and Tobago. Crop Sci 49:564–572
Kane N, Sveinsson S, Dempewolf H, Yang JY, Zhang D, Engels JMM, Cronk Q (2012) Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am J Bot 99:320–329
Kim S, Lee Y-P, Lim H, Ahn Y, Sung SK (2009) Identification of highly variable chloroplast sequences and development of cpDNA-based molecular markers that distinguish four cytoplasm types in radish (Raphanus sativus L.). Theor Appl Genet 119:189–198
Loor RG, Risterucci AM, Courtois B, Fouet O, Jeanneau M, Rosenquist E, Amores F, Vasco A, Medina M, Lanaud C (2009) Tracing the native ancestors of the modern Theobroma cacao L. population in Ecuador. Tree Genetics & Genomes 5:421–433
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079
Marshall HD, Newton C, Ritland K (2002) Chloroplast phylogeography and evolution of highly polymorphic microsatellites in lodgepole pine (Pinus contorta). Theor Appl Genet 104:367–378
Matsuo M, Ito Y, Yamauchi R, Obokata J (2005) The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast–nuclear DNA flux. The Plant Cell 17:665–675
Mogensen HL (1996) The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot 83:383–404
Motamayor JC, Risterucci AM, Lopez PA, Ortiz CF, Moreno A, Lanaud C (2002) Cacao domestication: I The origin of the cacao cultivated by the Mayas. Heredity 89:380–386
Motamayor JC, Risterucci AM, Heath M, Lanaud C (2003) Cacao domestication: II Progenitor germplasm of the Trinitario cacao cultivar. Heredity 91:322–330
Motilal LA, Zhang D, Umaharan P, Mischke S, Boccara M, Pinney S (2009) Increasing accuracy and throughput in large scale microsatellite fingerprinting of cacao field germplasm collections. Trop Plant Biol 2:23–37
Motilal LA, Zhang D, Umaharan P, Mischke S, Mooleedhar V, Meinhardt LW (2010) The relic Criollo cacao in Belize – genetic diversity and relationship with Trinitario and other cacao clones held in the International Cocoa Genebank Trinidad. Plant Genet Resource 8:106–115
Motilal LA, Zhang D, Umaharan P, Mischke S, Pinney S, Meinhardt LW (2011) Microsatellite fingerprinting in the International Cocoa Genebank, Trinidad: accession and plot homogeneity information for germplasm management. Plant Genet Resource 9:430–438
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Petit RJ, Vendramin GG (2007) Plant phylogeography based on organelle genes: an introduction. In: Weiss S, Ferrand N (eds) Phylogeography of Southern European refugia. Springer, Dordrecht, Netherlands, pp 23–97
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Pound FJ (1931) The genetic constitution of the cacao crop. First annual report on cacao research 1931. Government Printing Office, Port of Spain, Trinidad, pp 10–24
Pound FJ (1933) Criteria and methods of selection in cacao. In Second annual report on cacao research 1932. Government Printing Office, Port of Spain, Trinidad, pp 27–29
Pound FJ (1934) The progress of selection, 1933. In Third annual report on cacao research 1933. Government Printing Office, Port of Spain, Trinidad, pp 25–28
Pound FJ (1935) The progress of selection, 1934. In Fourth annual report on cacao research 1934. Government Printing Office, Port of Spain, Trinidad, pp 7–11
Pound FJ (1936) The progress of selection, 1935. In Fifth annual report on cacao research 1934. Government Printing Office, Port of Spain, Trinidad, pp 7–16
Powell W, Morgante M, Doyle JJ, McNicol JW, Tingey SV, Rafalski AJ (1996) Genepool variation in genus Glycine subgenus Soja revealed by polymorphic nuclear and chloroplast microsatellites. Genetics 144:793–803
Provan J, Corbett G, McNicol JW, Powell W (1997) Chloroplast DNA variability in wild and cultivated rice (Oryza spp.) revealed by polymorphic chloroplast simple sequence repeats. Genome 40:104–110
Provan J, Russell JR, Booth A, Powell W (1999) Polymorphic chloroplast simple sequence repeat primers for systematic and population studies in the genus Hordeum. Mol Ecol 8:505–511
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147
Rambaut A (2006–2009) FigTree. Tree figure drawing tool v.1.3.1, Institute of Evolutionary Biology, University of Edinburgh, UK
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, New Jersey, USA, pp 365–386
Shephard CY (1932) The cacao industry of Trinidad: some economic aspects, Part I. Government Printing office, Port of Spain, Trinidad, pp 95–100
Silva CRS, Albuquerque PSB, Ervedosa FR, Mota JWS, Figueira A, Sebbenn AM (2010) Understanding the genetic diversity, spatial genetic structure and mating system at the hierarchical levels of fruits and individuals of a continuous Theobroma cacao population from the Brazilian Amazon. Heredity 106:973–985
Small RL, Ryburn JA, Cronn RC, Seelanan T, Wendel JF (1998) The tortoise and the hare: choosing between noncoding plastome and nuclear ADH sequences for phylogeny reconstruction of a recently diverged plant group. Am J Bot 85:1301–1315
Sukumaran J, Holder MT (2010) DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571
Sveinsson S, Kane NC, Dempewolf H, Zhang D, Cronk QC (2010) Theobroma cacao chloroplast, complete genome. Website http://www.ncbi.nlm.nih.gov/nuccore/HQ244500 Accessed 15 January 2012
Takayama K, Kajita T, Murata J, Yoichi T (2006) Phylogeography and genetic structure of Hibiscus tiliaceus – speciation of a pantropical plant with sea-drifted seeds. Mol Ecol 15:2871–2881
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Toxopeus H (1985) Botany, types and populations. In: Wood GAR, Lass RA (eds) Cocoa, 4th edn. Longman, London, pp 11–37
Wendel JF (1989) New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci 86:4132–4136
Wood GAR (1985) History and development. In: Wood GAR, Lass RA (eds) Cocoa, 4th edn. Longman, London, pp 1–10
Yang JY, Motilal LA, Dempewolf H, Maharaj K, Cronk QC (2011) Chloroplast microsatellite primers for cacao (Theobroma cacao) and other Malvaceae. Am J Bot 98:e372–374
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Dissertation, University of Texas at Austin, Texas, USA
Acknowledgements
We thank the World Bank for a Development Marketplace grant awarded to QC, HD and Johannes Engels and coordinated by Bioversity International. The Staff of Cocoa Research, Centeno, Trinidad is highly acknowledged for identifying and collecting leaves from the farmers’ cacao trees, sometimes under very challenging conditions. We thank Chris Grassa for analytical assistance, and Brian Irish (USDA, ARS) and Kyle Wallick (USBG) for providing leaf samples. Stephen Pinney (USDA, ARS) and Kasey Gordon (CRU) are thanked for assistance with DNA extractions. Jon Armstrong and Jarret Glasscock (Cofactor Genomics, St. Louis) provided valuable sequencing assistance. We also gratefully acknowledge the laboratory support from an NSERC (Canada) discovery grant to Quentin Cronk.
Data Archiving Statement
Short-read sequences generated by the Illumina GA-II platform to assemble the T. cacao chloroplast genome have been deposited in the NIH Sequence Read Archive (SRA). The SRA accession number is SRA048198. The whole chloroplast genome sequences of the nine T. cacao genotypes and one T. grandiflorum genotype were deposited in GenBank. The accession numbers for these sequences are JQ228379-JQ228389. The DNA sequences used to obtain the ten SNPs were deposited in GenBank as well. The GenBank accession numbers for the first SNP primer pair are JX413598–JX413691; and the GenBank accession number for the second primer pair are JX413692–JX413781. The GenBank accession numbers for the SSR primers are JF979116–JF979124.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Sederoff
Rights and permissions
About this article
Cite this article
Yang, J.Y., Scascitelli, M., Motilal, L.A. et al. Complex origin of Trinitario-type Theobroma cacao (Malvaceae) from Trinidad and Tobago revealed using plastid genomics. Tree Genetics & Genomes 9, 829–840 (2013). https://doi.org/10.1007/s11295-013-0601-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-013-0601-4