Abstract
Compound-specific stable isotope analysis (CSIA) of amino acids is a new method that enables estimates of trophic position for consumers in food webs. We examined the nitrogen isotopic composition (δ15N) of amino acids of Japanese social insects (three bee, three wasp, and four hornet species) to evaluate the potential of CSIA of amino acids in studies of terrestrial food webs. For wasps, we also examined samples at different growth stages (ranging from egg to adult) to assess the effect of metamorphosis on CSIA estimates of trophic position. The δ15N values of bulk tissues for Japanese social insects are only weakly correlated with the biologically expected trophic positions. In contrast, the trophic positions estimated from the δ15N values of amino acids (yielding values of between 2.0 and 2.3 for bees, between 2.8 and 3.3 for wasps, and between 3.5 and 4.1 for hornets) are consistent with the biologically expected trophic positions for these insects (i.e., 2.0 for bees, 3.0 for wasps, and 3.0–4.0 for hornets). Although large variability is observed among the δ15N values of individual amino acids (e.g., ranging from 3.0 to 14.9‰ for phenylalanine), no significant change is observed in the trophic position during wasp metamorphosis. Thus, the CSIA of amino acids is a powerful tool for investigating not only aquatic food webs but also terrestrial food webs with predatory insects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Knowledge of the trophic position of organisms in food webs allows understanding of biomass flow and trophic linkages in complex networks of ecosystems. Compound-specific stable isotope analysis (CSIA) of amino acids is a new method that enables estimates of the trophic position of organisms in food webs (e.g., McClelland and Montoya 2002; Popp et al. 2007; Chikaraishi et al. 2009). It has been proposed that a comparison between large 15N-enrichment (+8.0‰) in glutamic acid and little change (+0.4‰) in phenylalanine, with each increase in trophic level, provides a precise estimate of the trophic position of organisms in food webs (Fig. 1). This approach is based on isotopic fractionations associated with the metabolic processes of two common amino acids: glutamic acid shows significant increase of δ15N values during reactions (e.g., transamination and deamination) that cleaves the carbon–nitrogen bond, whereas phenylalanine shows little change in δ15N values during conversion to tyrosine that neither forms nor cleaves the carbon–nitrogen bond (Chikaraishi et al. 2007). Chikaraishi et al. (2009, 2010a) established that the trophic position (TPGlu/Phe) of organisms could be estimated from the nitrogen isotopic compositions of glutamic acid (δ15NGlu) and phenylalanine (δ15NPhe), as follows:
where β represents the isotopic difference between glutamic acid and phenylalanine in primary producers (−3.4‰ for aquatic cyanobacteria and algae, +8.4‰ for terrestrial C3 plants, and −0.4‰ for terrestrial C4 plants; Fig. 1). This method has three key advantages. First, the trophic position is estimated based on the δ15N values of amino acids only from the target organism. Consequently, unlike bulk isotopic methods (i.e., determination of the trophic position from bulk nitrogen isotope analysis; e.g., Minagawa and Wada 1984), this method does not require characterization of the δ15N values of primary producers or baseline consumers. Second, small accuracy is expected; the standard deviation (1σ) of accuracy in the trophic position (=[actual TP] − [TPGlu/Phe]) is only 0.12 units for aquatic organisms (Chikaraishi et al. 2009) and 0.20 units for terrestrial organisms (Chikaraishi et al. 2010a). Third, a small sample size (nanomolar levels of nitrogen) is required for CSIA of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry (GC/C/IRMS) (Merritt and Hayes 1994; Chikaraishi et al. 2010b). As a result, CSIA of amino acids overcomes several of the challenges associated with bulk isotopic methods (e.g., O’Reilly et al. 2002; Post 2002). Consequently, the CSIA of amino acids has been used to elucidate the trophic position of organisms in recent ecological studies of shrimp in the sub-Antarctic archipelago (Pakhomov et al. 2004), krill in the Antarctic (Schmidt et al. 2004, 2006), plankton in the central Pacific (McCarthy et al. 2007), yellow-fin tuna in the eastern tropical Pacific (Popp et al. 2007), zooplankton near Hawaii (Hannides et al. 2009), and penguin in the Southern Ocean (Lorrain et al. 2009).
Recent evidence suggests that CSIA of amino acids is also useful in the evaluation of terrestrial food webs (Chikaraishi et al. 2010a; Naito et al. 2010; Styring et al. 2010). In particular, Chikaraishi et al. (2010a) demonstrated that a 15N enrichment factor of +7.6‰ (Eq. 1) describes the trophic relationship between caterpillars and plant leaves (i.e., diet), which is consistent with relationship found between aquatic organisms and their diets (Chikaraishi et al. 2009). However, few studies have used CSIA of amino acids to evaluate terrestrial food webs. In terrestrial ecosystems, insects are abundant, and are the dominant consumers at many trophic levels (e.g., Price et al. 1980). However, the factors controlling the isotopic composition of amino acids and the tropic position of insects remain poorly understood. In particular, many insects undergo a metamorphosis from larval to adult stages. Little information is available regarding isotopic and trophic changes associated with metamorphosis, from feeding during the larval and adult stages through starvation during the chrysalis stage. Also, pollen and nectar are basal resources in terrestrial food webs (e.g., Memmott 1999; O’Reilly et al. 2002, Patt et al. 2003), and feeding on pollen and nectar by pollinators is important for maintaining plant–pollinator interactions and terrestrial food webs (Memmott 1999). However, investigations of feeding on pollen and nectar would be difficult using only bulk δ15N values (Patt et al. 2003), because large variations exist among the δ15N values of these foods.
To further evaluate the applicability of CSIA of amino acids for food webs in terrestrial ecosystems, we examined the nitrogen isotopic compositions of amino acids in 45 samples of social insects, including three bee, three wasp, and four hornet species, as well as in 20 samples of reference materials including plant, caterpillar, aphid, and ladybug samples. For the three wasp species, we examined 26 samples, which included wasps at different growth stages (from egg to adult), which were used to evaluate the effect, if any, of metamorphosis on estimates of trophic position.
Materials and methods
All samples were collected from a farm near Yokohama, Japan (35°08′N, 139°07′E) (Table 1). This farm cultivates fruits (e.g., oranges and chestnuts) and vegetables (e.g., carrots, broccoli, eggplants, potatoes, radishes, and tomatoes), and all are C3 plants. Samples of adult bees (Apis mellifera, Bombus diversus diversus, and Xylocopa appendiculata circumvolans), wasps (Polistes japonicus japonicus, Polistes rothneyi iwatai, and Parapolybia indica), and hornets (Vespa ducalis pulchra, Vespa mandarinia japonica, Vespa simillima xanthoptera, and Vespula flaviceps lewisii) were collected from the farm. For wasps, we also collected different growth stages of samples: egg, larva, chrysalis, and wasps newly emerged from their nests. Leaves from several plants: Brassica oleracea (broccoli), Daucus carota (carrot), Castanea crenata (chestnut), Citrus unshiu (orange), Raphanus sativus (radish), Solanum lycopersicum (tomato), Solanum melongena (eggplant), and Solanum tuberosum (potato), nuts from C. crenata., and caterpillars, Pieris rapae, found on the leaves of B. oleracea were also collected as reference materials for primary producers and baseline consumers at this farm. Samples of the aphid Aphidoidea sp. and the ladybug Harmonia axyridis were also collected from the C. crenata leaves, as examples of insect samples with well-established trophic positions (i.e., 2.0 for aphids and 3.0 for ladybugs); aphids feed on the sap from C. crenata and ladybugs feed on the aphids.
Sample surfaces were washed with distilled water to remove contaminants. Samples were then freeze-dried and crushed to a fine powder. Powdered samples were stored at −20°C prior to analysis. The carbon and nitrogen isotopic compositions of the bulk sample materials were determined using a Thermo Fisher Scientific Flash EA (EA1112) coupled to a DeltaplusXP IRMS with a ConFlo III interface (Ogawa et al. 2010). Carbon and nitrogen isotopic compositions are reported in the standard delta (δ) notation relative to the Vienna Pee Dee Belemnite standard (VPDB) and to atmospheric nitrogen (Air), respectively.
All samples were prepared for CSIA of amino acids with HCl hydrolysis followed by N-pivaloyl/isopropyl (Pv/iPr) derivatization, according to the methods described by Chikaraishi et al. (2007). In brief, samples were hydrolyzed using 12 M HCl at 100°C. The hydrolysate was washed with n-hexane/dichloromethane (6:5, v/v) to remove hydrophobic constituents (e.g., lipids). Then, derivatizations were performed sequentially with thionyl chloride/2-propanol (1:4, v/v) and pivaloyl chloride/dichloromethane (1:4, v/v). The Pv/iPr derivatives of the amino acids were extracted with n-hexane/dichloromethane (6:5, v/v). The nitrogen isotopic compositions of individual amino acids were determined by GC/C/IRMS using an Agilent Technologies 6890N GC coupled to a Thermo Fisher Scientific DeltaplusXP IRMS with a GC-C/TC III interface. The analytical conditions for the GC/C/IRMS analyses are described in detail by Chikaraishi et al. (2008, 2010a). Standard mixtures of ten amino acids (alanine, glycine, valine, leucine, norleucine, aspartic acid, serine, glutamic acid, phenylalanine, and hydro) with known δ15N values (ranging from −7.3 to +22.7‰) were analyzed after every three to four samples during GC/C/IRMS analytical sessions to assess the reproducibility of the isotope measurements and to normalize the δ15N values of amino acids in samples. Three pulses of reference N2 gas (−4.3‰) were discharged into the IRMS at the beginning and end of each chromatogram for both standard mixtures and samples. The isotopic compositions of amino acids in samples are expressed relative to atmospheric N2 on scales normalized to the known δ15N values of standard amino acids.
Nitrogen isotopic compositions (δ15N) were determined for the following ten amino acids in samples: alanine, glycine, valine, leucine, isoleucine, proline, serine, methionine, glutamic acid, and phenylalanine. These amino acids were chosen because their peaks were always well separated with baseline resolution in the chromatogram (Metges et al. 1996; Chikaraishi et al. 2010b). The accuracy obtained for the standards and samples was always better than ±0.5‰ for sample sizes of ≥30 ng N. Glutamine was converted to glutamic acid during acid hydrolysis; as a result, the α-amino group of glutamine contributed to the δ15N value calculated for glutamic acid. Additional amino acids were not included in the study due to difficulties associated with sample preparation; primarily, the co-elution of a portion of aspartic acid with threonine during chromatography and the absence of arginine, cysteine, histidine, lysine, tyrosine, and tryptophan from chromatograms, which is attributed to the decomposition of samples during preparation or low sample recovery (Chikaraishi et al. 2010b). The trophic position of samples was calculated from the δ15N values for glutamic acid and phenylalanine using Eq. (1) and a β value of +8.4‰.
Results and discussion
δ13C and δ15N values of bulk tissues
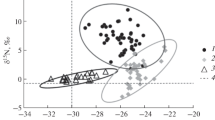
The carbon and nitrogen isotopic compositions determined for the bulk tissues of the bees, wasps, and hornets are listed in Table 1, together with those of reference materials (plants, caterpillars, aphids, and ladybugs) collected from the same farm. The δ13C values of bulk tissues are between −26.8 and −25.0‰ for bees, −30.1 and −26.1‰ for wasps, and −25.8 and –24.6‰ for hornets. These δ13C values are similar to or slightly more positive than those determined for plants (between −32.5 and −27.7‰), caterpillars (between −29.6 and −26.9‰), aphids (−21.8), and ladybugs (between −29.0 and −26.5‰). These results are consistent with the previously observed δ13C shift in food webs (−0.6 to +2.7‰ at each trophic level; e.g., DeNiro and Epstein 1981). Thus, the examined insects are presumed to feed entirely on plant materials or consumers in C3 food webs.
Field observations reveal that bee species (i.e., A. mellifera, B. diversus, and X. appendiculata) feed on nectar and pollen from flowers; wasp species (i.e., P. indica, P. japonicus, and P. rothneyi) feed mainly on caterpillars found on plant leaves; the hornet species V. ducalis feeds solely on wasps; and other hornet species (i.e., V. flaviceps, V. mandarinia, and V. simillima) are carnivorous predators that feed on various insects, such as caterpillars, bees, and wasps (e.g., Takamizawa 2005). Therefore, the biologically expected trophic positions are 2.0 for bees, 3.0 for wasps, 4.0 for the hornet species V. ducalis, and between 3.0 and 4.0 for the other hornet species. The δ15N values of bulk tissues are between +1.6 and +5.1‰ for bees, between +1.1 and +7.2‰ for wasps, and between +4.5 and +5.6‰ for hornets, which largely overlaps the range of δ15N values for plant materials (between −2.8 and +5.9‰). If an average δ15N value for the examined plant materials is simply employed as a δ15N baseline for the determination of trophic position in the bulk method, trophic positions may be estimated from the δ15N values of bulk tissues (i.e., between 0.5 and 1.5 for bees, between 0.3 and 2.1 for wasps, and between 1.3 and 1.7 for hornets; see Table 1). The estimated trophic positions show only a weak correlation with the expected trophic positions for these insects.
The weak correlation observed between estimated and expected trophic positions is largely attributed to the large variation among the δ15N values exhibited by plants in modern terrestrial environments, especially in the case of a farm. This large variation among δ15N values may also reflect the influence of human activities on terrestrial environments (e.g., Denton et al. 2001; Commisso and Nelson 2006). In fact, in our study, significant variation in δ15N values (ranging from −2.8 to +5.9‰) was observed for bulk plant leaves. This range is equivalent to approximately 2.6 times the 15N-enrichment factor in the bulk method (i.e., 3.4‰; Minagawa and Wada 1984).
Accuracy in CSIA of amino acids
The nitrogen isotopic compositions of ten amino acids in terrestrial insects (bees, wasps, and hornets) and the reference materials (plants, caterpillars, aphids, and ladybugs) are given in Table 1. As with the bulk isotopic compositions, large variability is observed among the δ15N values of amino acids; for phenylalanine, the δ15N values are only weakly increased by trophic level, yet the δ15N values are still highly variable (between +5.7 and +12.9‰ for bees, +3.0 and +14.9‰ for wasps, and +7.7 and +9.7‰ for hornets), likely reflecting large variation among the δ15N values of plants (i.e., the food web base) at this farm.
When CSIA of amino acids is used for plants and insects with well-established trophic positions, the trophic positions estimated from the δ15N values of glutamic acid and phenylalanine are between 1.0 and 1.3 for plants, between 1.8 and 2.2 for aphids, and 3.1 for ladybugs (Fig. 2). The potential uncertainly in the trophic position calculated by taking into account the propagation of uncertainty (i.e., 0.5‰ for δ15NGlu and δ15NPhe, 1.6‰ for β, and 1.2‰ for denominator: Chikaraishi et al. 2010a, b) in Eq. (1) is only 0.23 for plants, 0.28 for aphids, and 0.40 for ladybugs. Thus the trophic positions estimated by CSIA of amino acids are consistent with the actual trophic positions for these plants and insects (1.0 for plants, 2.0 for aphides, and 3.0 for ladybugs). The standard deviation (1σ) on the accuracy (=[actual TP] − [TPGlu/Phe]) is 0.11 units for these plants and insects, which is somewhat smaller than that (1σ = 0.20) estimated from plants and caterpillars in our previous study (Chikaraishi et al. 2010a). Combined with the dataset for plants and caterpillars provided by Chikaraishi et al. (2010a), here we employ 0.17 unit (1σ) as the accuracy in the trophic position estimated by CSIA of amino acids for terrestrial C3 food webs.
Nitrogen isotopic compositions of glutamic acid (filled circles) and phenylalanine (open circles), and trophic positions, estimated by the amino acid method (TPGlu/Phe, filled squares), for plants (C. crenata), aphids (Aphidea sp.), and ladybugs (H. axyridis). Bars represent potential uncertainly in TPGlu/Phe calculated by taking into account the propagation of 1σ for δ15NGlu, δ15NPhe, β, and denominator (7.6) in Eq. (1)
Based on triplicate analysis for representative species, the standard deviation (1σ) for the comparison of trophic positions estimated by CSIA of amino acids is <0.21 (0.13 on average) units for all samples (i.e., plant leaves, caterpillars, bees, wasps and hornets) (Fig. 3a). This variation is almost identical or smaller than the accuracy (1σ = 0.17) of this method. The insects of interest may exhibit large variation among the bulk δ15N values of different body tissues (e.g., leg and wing) because of highly variable compositions of nitrogen-containing molecules, which reflect their feeding habits and life strategies. In fact, the bulk δ15N values for different body tissues of a wasp (P. rothneyi #15) exhibit a spread of 2.4‰ among abdomen (+3.8‰), leg and wing (+5.4‰), thorax (+5.7‰), and head (+6.4‰) tissues (Table 1). However, as shown in Fig. 3b, the δ15N values of amino acids, particularly those of glutamic acid and phenylalanine, are within error among tissues (i.e., within 1σ of 0.4‰ for the comparison of the δ15N values). Moreover, the estimated trophic positions are consistent for different tissues (i.e., within 1σ of 0.03 units for the comparison of the TPGlu/Phe values). Thus, no substantial difference is observed in both the δ15N values of amino acids and the estimated trophic positions among different body tissues; e.g., leg and wing.
Nitrogen isotopic compositions of glutamic acid (filled circles) and phenylalanine (open circles), and trophic positions, estimated by the amino acid method (TPGlu/Phe, filled squares), for a plant (B. oleracea), caterpillar (P. rapae), bee (A. mellifera), wasp (P. rothneyi), and hornet (V. ducalis) samples (analyzed in triplicate), and b different body tissues of a wasp (P. rothneyi). Bars represent potential uncertainly in TPGlu/Phe calculated by taking into account the propagation of 1σ for δ15NGlu, δ15NPhe, β, and denominator (7.6) in Eq. (1)
Effect of metamorphosis
The lifestyle (e.g., feeding habits) of many insects shows a marked change during metamorphosis from larva to adult. For wasps, after the egg hatches, they feed mainly on caterpillars (hunted by adult worker wasps) during their larva stage, and do not feed during metamorphose in cocoons during their chrysalis stage (e.g., Takamizawa 2005). In a previous study on the Diptera Chironomus acerbiphilus (Doi et al. 2007), metamorphosis resulted in an increase of between 0.7 and 1.6‰ in δ15N values of bulk tissues from larva to adult. Similar increases in δ15N values have been observed for Lepidoptera (McCutchan et al. 2003; Tabbets et al. 2008) and Neuroptera (Patt et al. 2003).
However, as shown in Fig, 4, our results demonstrate that for wasps (P. indica, P japonicus, and P. rothney), trophic position does not change significantly (1σ < 0.14 units for the comparison of the TPGlu/Phe values) from the egg to adult stages, despite marked differences among the δ15N values for wasps at different growth stages (e.g., for phenylalanine, between +3.0 and +11.2‰ for P indica, between +8.7 and +14.1‰ for P japonicus, and between +8.3 and +14.9‰ for P rothneyi). These results suggest that amino acids are not metabolized as energy sources during wasp metamorphosis in cocoons (it could be considered as a semi-closed system), and that metamorphosis has a negligible effect on the δ15N values of amino acids and the trophic positions of wasps. Rather, the observed variations in δ15N values for both bulk tissues and amino acids may reflect changes (or variations) in the δ15N values of their diet as they feed during the larva stage.
Nitrogen isotopic compositions of glutamic acid (filled circles) and phenylalanine (open circles), and trophic positions, estimated by the amino acid method (TPGlu/Phe, filled squares), for three wasps at various growth stages. Bars represent potential uncertainly in TPGlu/Phe calculated by taking into account the propagation of 1σ for δ15NGlu, δ15NPhe, β, and denominator (7.6) in Eq. (1)
Trophic positions of bees, wasps, and hornets
The bulk δ15N values vary significantly for plants (between −2.8 and +5.9‰) and insects (between +1.1 and +7.2‰) collected from the farm. Trophic positions estimated by the bulk method have only a weak correlation with the biologically expected trophic positions of these insects (Table 1). However, the trophic positions estimated by CSIA of amino acids are 2.0 and 2.1 for A. mellifera, 2.3 for B. diversus, 2.1 for X. appendiculata, between 2.8 and 3.1 for P. indica, between 2.9 and 3.2 for P. japonicus, and between 2.8 and 3.3 for P. rothneyi (Fig. 5). Considering the accuracy associated with this method (1σ = 0.17), the estimated trophic positions are well consistent with the biologically expected trophic positions of the bees that feed on nectar and pollen from flowers and of the wasps that feed primarily on caterpillars found on plant leaves. Moreover, the trophic positions estimated for hornets by CSIA of amino acids (between 3.9 and 4.0 for V. ducalis and 3.5 for the other hornets, i.e., V. flaviceps, V. mandarinia, and V. simillima; Fig. 5), are consistent with the biologically expected trophic positions for both hornets that feed solely on wasps and other hornets that feed on various insects (e.g., caterpillars, bees, and wasps), respectively.
Herbivorous insects feed on various types of plant tissues such as leaves, twigs, bark, and nectar. In the present study, bees feed on nectar and pollen, caterpillars feed on leaves, and aphids feed on sap. The trophic positions of these insects are estimated to be between 1.8 and 2.3 (2.1 on average), which are reasonable values for the herbivores. Our study suggests that variation among food sources (i.e., plant tissues) does not significantly affect estimates of trophic positions for herbivores. Adult wasps feed on the excretion of larvae as well as nectars from flowers (e.g., Takamizawa 2005), as a result, the trophic positions of adult wasps may be expected to be different from those of larvae and chrysalis. However, no substantial difference is observed among the trophic positions of wasps at different growth stages (i.e., egg, larva, chrysalis, and even adult). This indicates that feeding on excretions and nectars during the adult stages has little effect on the trophic position of adult wasps.
The trophic position of omnivore hornets (V. flaviceps, V. mandarinia, and V. simillima) is estimated to be 3.5. Bulk δ15N values have been used to estimate trophic positions; however, this is particularly difficult in the case of omnivores. If an omnivore assimilates various food sources with different C:N ratios (i.e., plant and animal), analysis with a simple model using bulk δ15N values would fail to incorporate differences in C:N stoichiometry among food sources (Post 2002). Using CSIA of amino acids, the trophic position of an omnivore may be estimated using a simple model that considers differences in food quality, because amino acids are assimilated independently of the elemental stoichiometry.
Conclusions
We evaluated the potential of CSIA of amino acids for estimating the trophic position of terrestrial insects (i.e., bees, wasps, and hornets) in a case study of terrestrial food webs. The CSIA estimated trophic positions (yielding values of between 2.0 and 2.3 for bees, between 2.8 and 3.3 for wasps, and between 3.5 and 4.1 for hornets) are well consistent with the biologically expected trophic positions of these insects (i.e., 2.0 for bees, 3.0 for wasps, and 3.0–4.0 for hornets) given the accuracy associated with this method (1σ = 0.17). In addition, we determined that wasp metamorphosis may not significantly affect the δ15N values of amino acids as well as trophic position. Thus, estimates of trophic position based on CSIA of amino acids are useful in investigations of terrestrial food webs with predatory insects.
Our results suggest that CSIA of amino acids is a useful tool for assessing the structure of food webs and biogeochemical processes in both aquatic and terrestrial environments. However, the present method cannot be applied directly to a more complex food web, where the basal resources include terrestrial C3 and C4 plants, as well as aquatic plants, because the factor β in Eq. (1) is different among the basal resources (Fig. 1). In such cases, the mixing ratio among different resources should be quantified by other proxies prior to estimating trophic position. We suggest the use of carbon isotope analysis of bulk tissues or amino acids to address this problem. Advances in this field are expected from recent developments and improvements in methodologies for CSIA of amino acids using δ13C (Corr et al. 2007a, b; Smith et al. 2009; Chikaraishi and Ohkouchi 2010; Choy et al. 2010). CSIA of amino acids, using both δ13C and δ15N values, would facilitate the study of ecological food webs where C3, C4, and aquatic plants are present.
References
Chikaraishi Y, Ohkouchi N (2010) An improved method for precise determination of carbon isotopic composition of amino acids. In: Ohokouchi N, Tayasu I, Koba K (eds) Earth, life, and isotopes. Kyoto University Press, Kyoto, pp 355–366
Chikaraishi Y, Kashiyama Y, Ogawa NO, Kitazato H, Ohkouchi N (2007) Biosynthetic and metabolic controls of nitrogen isotopic composition of amino acids in marine macroalgae and gastropods: implications for aquatic food web studies. Mar Ecol Prog Ser 342:85–90
Chikaraishi Y, Kashiyama Y, Ogawa NO, Kitazato H, Satoh M, Nomoto S, Ohkouchi N (2008) A compound-specific isotope method for measuring the stable nitrogen isotopic composition of tetrapyrroles. Org Geochem 39:510–520
Chikaraishi Y, Ogawa NO, Kashiyama Y, Takano Y, Suga H, Tomitani A, Miyashita H, Kitazato H, Ohkouchi N (2009) Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol Oceanogr Meth 7:740–750
Chikaraishi Y, Ogawa NO, Ohkouchi N (2010a) Further evaluation of the trophic level estimation based on nitrogen isotopic composition of amino acids. In: Ohokouchi N, Tayasu I, Koba K (eds) Earth, life, and isotopes. Kyoto University Press, Kyoto, pp 37–51
Chikaraishi Y, Takano Y, Ogawa NO, Ohkouchi N (2010b) Instrumental optimization for compound-specific nitrogen isotope analysis of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry. In: Ohokouchi N, Tayasu I, Koba K (eds) Earth, life, and isotopes. Kyoto University Press, Kyoto, pp 367–386
Choy K, Smith CI, Fuller BT, Richards MP (2010) Investigation of amino acid δ13C signatures in bone collagen to reconstruct human palaeodiets using liquid chromatography–isotope ratio mass spectrometry. Geochim Cosumochim Acta 74:6093–6111
Commisso RG, Nelson DE (2006) Modern plant δ15N values reflect ancient human activity. J Archaeol Sci 33:1167–1176
Corr LT, Berstan R, Evershed RP (2007a) Optimisation of derivatisation procedures for the determination of δ13C values of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry. Rapid Commun Mass Spectrom 21:3759–3771
Corr LT, Berstan R, Evershed RP (2007b) Development of N-acetyl methyl ester derivatives for the determination of δ13C values of amino acids using gas chromatography-combustion-isotope ratio mass spectrometry. Anal Chem 79:9082–9090
DeNiro MJ, Epstein S (1981) Influence of diet on the distribution of nitrogen isotopes in animals. Geochim Cosmochim Acta 45:341–351
Denton TM, Schmidt S, Critchley C, Stewart GR (2001) Natural abundance of stable carbon and nitrogen isotopes in Cannabis sativa reflects growth conditions. Aust J Plant Physiol 28:1005–1012
Doi H, Kikuchi E, Takagi S, Shikano S (2007) Changes in carbon and nitrogen stable isotopes of chironomid larvae during growth, starvation and metamorphosis. Rapid Commun Mass Spectrom 21:997–1002
Hannides CCS, Popp BN, Landry MR, Graham BS (2009) Quantification of zooplankton trophic position in the North Pacific Subtropical Gytr using stable nitrogen isotopes. Limnol Oceanogr 54:50–61
Lorrain A, Graham B, Ménard F, Popp B, Bouillon S, van Breugel P, Cherel Y (2009) Nitrogen and carbon isotope values of individual amino acids: a tool to study foraging ecology of penguins in the Southern Ocean. Mar Ecol Prog Ser 391:293–306
McCarthy MD, Benner R, Lee C, Fogel ML (2007) Amino acid nitrogen isotopic fractionation patterns as indicators of heterotrophy in plankton, particulate, and dissolved organic matter. Geochim Cosmochim Acta 71:4727–4744
McClelland JW, Montoya JP (2002) Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 83:2173–2180
McCutchan JM Jr, Lewis WM Jr, McGrath CC (2003) Variation in trophic shift for stable isotope ratios of carbon, nitrogen, and sulfur. OIKOS 102:378–390
Memmott J (1999) The structure of a plant-pollinator food web. Ecol Lett 2:276–280
Merritt DA, Hayes JM (1994) Nitrogen isotopic analyses by isotope-ratio-monitoring gas chromatography/mass spectrometry. J Am Soc Mass Spectrom 5:387–397
Metges CC, Petzke KJ, Henning U (1996) Gas chromatography/combustion/isotope ratio mass spectrometric comparison of N-acetyl- and N-pivaloyl amino acid esters to measure 15N isotopic abundances in physiological samples: a pilot study on amino acid synthesis in the upper gastro-intestinal tract of minipigs. J Mass Spectrom 31:367–376
Minagawa M, Wada E (1984) Stepwise enrichment of 15N along food chains: further evidences and the relation between δ15N and animal age. Geochim Cosmochim Acta 48:1135–1140
Naito YI, Honch NV, Chikaraishi Y, Ohkouchi N, Yoneda M (2010) Quantitative evaluation of marine protein contribution in ancient diets based on nitrogen isotope ratios of individual amino acids in bone collagen: an investigation at the Kitakogane Jomon site. Am J Phys Anthropol J 143:31–40
O’Reilly CM, Hecky RE, Cohen AS, Plisnier P-D (2002) Interpreting stable isotopes in food webs: recognizing the role of time averaging at different trophic levels. Limnol Oceanogr 47:306–309
Ogawa NO, Nagata T, Kitazato H, Ohkouchi N (2010) Ultra sensitive elemental analyzer/isotope ratio mass spectrometer for stable nitrogen and carbon isotope analyses. In: Ohokouchi N, Tayasu I, Koba K (eds) Earth, life, and isotopes. Kyoto University Press, Kyoto, pp 339–353
Pakhomov EA, McClelland JW, Bernard K, Kaehler S, Montoya JP (2004) Spatial and temporal shifts in stable isotope values of the bottom-dwelling shrimp Nauticaris marionis at the sub-Antarctic archipelago. Mar Biol 144:317–325
Patt JM, Waunright SC, Hamilton GC, Whittinghill D, Bosley K, Dietrck J, Lashomb JH (2003) Assimilation of carbon and nitrogen from pollen and nectar by a predaceous larva and its effects on growth and development. Ecol Entomol 28:717–728
Popp BN, Graham BS, Olson RJ, Hannides CCS, Lott M, López-Ibarra G, Galván-Magaña F (2007) Insight into the trophic ecology of yellowfin tuna, Thunnus albacares, from compound-specific nitrogen isotope analysis of proteinaceous amino acids. In: Dawson TE, Siegwolf RTW (eds) Stable isotopes as indicators of ecological change. Academic Press, Dublin, pp 173–190
Post DM (2002) Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology 83:703–718
Price PW, Bouton CE, Gross P, McPheron BA, Thompson JN, Weis AE (1980) Interactions among three trophic levels: influence of plants on interactions between insect herbivores and natural enemies. Ann Rev Ecol Sys 11:41–65
Schmidt K, McClelland JM, Mente E, Montoya JP, Atkinson A, Voss M (2004) Trophic-level interpretation based on δ15N values: implication of tissue-specific fractionation and amino acid composition. Mar Ecol Prog Ser 266:43–58
Schmidt K, Atkinson A, Petzke K-J, Voss M, Pond DW (2006) Protozoans as a food source for Antarctic krill, Euphausia superba: complementary insights from stomach content, fatty acids, and stable isotopes. Limnol Oceanogr 51:2409–2427
Smith CI, Fuller BT, Choy K, Richards MP (2009) A three-phase liquid chromatographic method for δ13C analysis of amino acids from biological protein hydrolysates using liquid chromatography–isotope ratio mass spectrometry. Anal Biochem 390:165–172
Styring AK, Sealy JC, Evershed RP (2010) Resolving the bulk δ15N values of ancient human and animal bone collagen via compound-specific nitrogen isotope analysis of constituent amino acids. Geochim Cosmochim Acta 74:241–251
Tabbets TM, Wheeless LA, Marthinez del Rio C (2008) Isotopic enrichment without change in diet: an ontogenetic shift in δ15N during insect metamorphosis. Func Ecol 22:109–113
Takamizawa K (2005) The Japanese social wasps and bees. The Shinano Mainichi Shimbun, Nagano
Acknowledgments
We thank Mr. Mikio Chikaraishi (Yamani farm) for providing samples and Ms. Rutsu Hirono for drawing illustrations of insects. We are grateful to Dr. Yoshinori Takano (Biogeos, JAMSTEC) and Prof. Brian N. Popp (University of Hawaii) for expert advice and constructive discussion. We also thank handling editors and two anonymous reviewers for their valuable suggestions and thoughtful comments that are very helpful for improving the manuscript. This work was supported by Grant-in-Aid for Scientific Research of the JSPS (Y.C., N.O.O., H.D. and N.O.), and Grant-in-Aid for Creative Scientific Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Chikaraishi, Y., Ogawa, N.O., Doi, H. et al. 15N/14N ratios of amino acids as a tool for studying terrestrial food webs: a case study of terrestrial insects (bees, wasps, and hornets). Ecol Res 26, 835–844 (2011). https://doi.org/10.1007/s11284-011-0844-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11284-011-0844-1