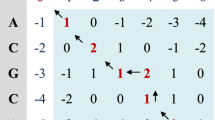
Abstract
Several problems in computational biology require the all-against-all pairwise comparisons of tens of thousands of individual biological sequences. Each such comparison can be performed with the well-known Needleman-Wunsch alignment algorithm. However, with the rapid growth of biological databases, performing all possible comparisons with this algorithm in serial becomes extremely time-consuming. The massive computational power of graphics processing units (GPUs) makes them an appealing choice for accelerating these computations. As such, CPU-GPU clusters can enable all-against-all comparisons on large datasets. In this work, we present four GPU implementations for large-scale pairwise sequence alignment: TiledDScan-mNW, DScan-mNW, RScan-mNW and LazyRScan-mNW. The proposed GPU kernels exhibit different parallelization patterns: we discuss how each parallelization strategy affects the memory accesses and the utilization of the underlying GPU hardware. We evaluate our implementations on a variety of low- and high-end GPUs with different compute capabilities. Our results show that all the proposed solutions outperform the existing open-source implementation from the Rodinia Benchmark Suite, and LazyRScan-mNW is the preferred solution for applications that require performing the trace-back operation only on a subset of the considered sequence pairs (for example, the pairs whose alignment score exceeds a predefined threshold). Finally, we discuss the integration of the proposed GPU kernels into a hybrid MPI-CUDA framework for deployment on CPU-GPU clusters. In particular, our proposed distributed design targets both homogeneous and heterogeneous clusters with nodes that differ amongst themselves in their hardware configuration.
















Similar content being viewed by others
References
Needleman, S. B., & Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology, 48, 443–453.
Smith, T. F., & Waterman, M. S. (1981). Identification of common molecular subsequences. Journal of Molecular Biology, 147(1), 195–197.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Hillis, D. M., Moritz, C., & Mable, B. K. (1996). Molecular systematics (2nd ed.). Sunderland: Sinauer Associates.
Nei, M., & Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular Biology and Evolution, 3(5), 418–426.
Pearson, W. R., & Lipman, D. J. (1988). Improved tools for biological sequence comparison. Proceedings of the National Academy of Sciences of the United States of America, 85(8), 2444–2448.
Altschul, S. F., Gish, W., Miller, W., et al. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
Altschul, S. F., Madden, T. L., Schaffer, A. A., et al. (1997). Gapped blast and Psi-blast : a new-generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402.
Li, H., Ruan, J., & Durbin, R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Research, 18(11), 1851–1858.
Langmead, B., Trapnell, C., Pop, M., et al. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 10(3), R25.
Myers, G. (1999). A fast bit-vector algorithm for approximate string matching based on dynamic programming. Journal of the ACM (JACM), 46(3), 395–415.
Benson, D. A., Cavanaugh, M., Clark, K., et al. (2013). GenBank. Nucleic Acids Research, 41(Database issue), D36–D42.
Meusemann, K., von Reumont, B. M., Simon, S., et al. (2010). A phylogenomic approach to resolve the arthropod tree of life. Molecular Biology and Evolution, 27(11), 2451–2464.
Pace, N. R. (2009). Mapping the tree of life: progress and prospects. Microbiology and Molecular Biology Reviews, 73(4), 565–576.
Parfrey, L. W., Grant, J., Tekle, Y. I., et al. (2010). Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Systems Biology, 59(5), 518–533.
Beja, O., Suzuki, M. T., Heidelberg, J. F., et al. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature, 415(6872), 630–633.
Kim, M., Morrison, M., & Yu, Z. (2011). Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiology Ecology, 76(1), 49–63.
Tringe, S. G., & Rubin, E. M. (2005). Metagenomics: DNA sequencing of environmental samples. Nature Reviews Genetics, 6(11), 805–814.
Venter, J. C., Remington, K., Heidelberg, J. F., et al. (2004). Environmental genome shotgun sequencing of the sargasso Sea. Science, 304(5667), 66–74.
Whitford, M. F., Forster, R. J., Beard, C. E., et al. (1998). Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe, 4(3), 153–163.
Cole, J. R., Wang, Q., Cardenas, E., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research, 37(Database issue), D141–D145.
Tarditi, D., Puri, S., & Oglesby, J. (2006). Accelerator: using data parallelism to program GPUs for general-purpose uses. SIGARCH Comput. Archit. News, 34(5), 325–335.
Che, S., Boyer, M., Meng, J., et al. (2009). “Rodinia: A benchmark suite for heterogeneous computing,” in Proc. of IISWC, pp. 44–54.
“Nvidia Applications Catalog” http://www.nvidia.com/docs/IO/123576/nv-applications-catalog-lowres.pdf
Vouzis, P. D., & Sahinidis, N. V. (2010). GPU-BLAST: using graphics processors to accelerate protein sequence alignment. Bioinformatics, 27(2), 182–188.
Schatz, M. C., Trapnell, C., Delcher, A. L., et al. (2007). High-throughput sequence alignment using graphics processing units. BMC Bioinformatics, 8, 474.
Walters, J. P., Meng, X., Chaudhary, V., et al. (2007). MPI-HMMER-boost: distributed FPGA acceleration. The Journal of VLSI Signla Processing Systems for Signal, Image, and Video Technology, 48(3), 6.
Pang, B., Zhao, N., Becchi, M., et al. (2012). Accelerating large-scale protein structure alignments with graphics processing units. BMC Res Notes, 5, 116.
Manavski, S. A., & Valle, G. (2008). CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment. BMC Bioinformatics, 9(Suppl 2), S10.
Liu, W., Schmidt, B., Voss, G., et al. (2007). Streaming algorithms for biological sequence alignment on GPUs. IEEE Transactions on Parallel and Distributed Systems, 19, 1270–1281.
Gao, Y., and Bakos, J. D. (2012). “GPU Acceleration of Pyrosequencing Noise Removal,” in Proc. of SAAHPC, Argonne, IL USA, pp. 94–101.
Liu, Y., Maskell, D. L., & Schmidt, B., (2009). “CUDASW++: Optimizing Smith-Waterman Sequence Database Searches for CUDA-enabled Graphics Processing Units,” BMC Research Notes, vol. 2, no. 73.
Wirawan, A., Kwoh, C. K., Hieu, N. T., et al. (2008). CBESW: sequence alignment on the playstation 3. BMC Bioinformatics, 9, 377.
Szalkowski, A., Ledergerber, C., Krahenbuhl, P., et al. (2008). SWPS3 - Fast multi-threaded vectorized Smith-Waterman for IBM cell/B.E. And x86/SSE2. BMC Res Notes, 1, 107.
Li, J., Ranka, S., & Sahni, S., (2012).“Pairwise sequence alignment for very long sequences on GPUs,” in Proc. of ICCABS, pp. 1–6.
Li, K.-B. (2003). ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics, 19(12), 2.
Biegert, A., Mayer, C., Remmert, M., et al. (2006). The MPI bioinformatics toolkit for protein sequence analysis. Nucleic Acids Research, 34, 5.
Henikoff, S., & Henikoff, J. G. (1992). Amino-acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences, U.S.A, 22, 10915–10919.
Hirschberg, D. S. (1975). A linear space algorithm for computing maximal common subsequences. Communications of the ACM, 18(6), 341–343.
Myers, E. W., & Miller, W. (1988). Optimal alignments in linear space. Computer applications in the biosciences: CABIOS, 4(1), 11–17.
Sanders, J., & Jabdrot, E., (2010). CUDA by Example: An Introduction to General-Purpose GPU Programming: Addison-Wesley Professional.
Acknowledgments
We thank the reviewers for their feedback. This work has been supported by NSF award CNS-1216756 and by equipment donations from Nvidia Corporation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Truong, H., Li, D., Sajjapongse, K. et al. Large-Scale Pairwise Alignments on GPU Clusters: Exploring the Implementation Space. J Sign Process Syst 77, 131–149 (2014). https://doi.org/10.1007/s11265-014-0883-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11265-014-0883-2