Abstract
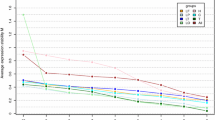
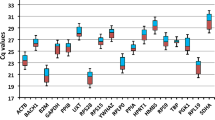
The common marmoset (Callithrix jacchus) is increasingly being used as a non-human primate animal model in biomedical research. To perform accurate quantitative analysis of gene expression by quantitative reverse transcription polymerase chain reaction, reliable reference genes should be selected. In this study, we evaluated the expressions of 11 widely used reference genes: ACTB, ATP5F1, B2M, GAPDH, HPRT1, PGK1, PPIA, RN18S1, RPLP0, TBP and UBC in 12 tissues and five brain areas of healthy common marmosets. NormFinder and geNorm indicated that the most suitable reference genes for cross-sectional studies of the 17 tissues were RN18S1 and RPLP0. Conversely, ACTB and PPIA were the most suitable for analyzing brain samples; however, the expression of PGK1 fluctuated among brain areas. These results indicate that suitable reference genes differ between the tissues examined. This study provides fundamental information for gene expression studies of the common marmoset and highlights the importance of validating reference genes before quantification of target mRNAs.


Similar content being viewed by others
References
Mansfield K (2003) Marmoset models commonly used in biomedical research. Comp Med 53(4):383–392
Baker HF, Ridley RM, Duchen LW, Crow TJ, Bruton CJ (1993) Experimental induction of beta-amyloid plaques and cerebral angiopathy in primates. Ann N Y Acad Sci 695:228–231
Maclean CJ, Baker HF, Ridley RM, Mori H (2000) Naturally occurring and experimentally induced beta-amyloid deposits in the brains of marmosets (Callithrix jacchus). J Neural Transm 107(7):799–814
Gnanalingham KK, Smith LA, Hunter AJ, Jenner P, Marsden CD (1993) Alterations in striatal and extrastriatal D-1 and D-2 dopamine receptors in the MPTP-treated common marmoset: an autoradiographic study. Synapse 14(2):184–194
Roeling TA, Docter GJ, Voorn P, Melchers BP, Wolters EC, Groenewegen HJ (1995) Effects of unilateral 6-hydroxydopamine lesions on neuropeptide immunoreactivity in the basal ganglia of the common marmoset, Callithrix jacchus, a quantitative immunohistochemical analysis. J Chem Neuroanat 9(3):155–164
Kendall AL, Rayment FD, Torres EM, Baker HF, Ridley RM, Dunnett SB (1998) Functional integration of striatal allografts in a primate model of Huntington’s disease. Nat Med 4(6):727–729
t Hart BA, van Meurs M, Brok HP, Massacesi L, Bauer J, Boon L, Bontrop RE, Laman JD (2000) A new primate model for multiple sclerosis in the common marmoset. Immunol Today 21(6):290–297
Barros M, Tomaz C (2002) Non-human primate models for investigating fear and anxiety. Neurosci Biobehav Rev 26(2):187–201
Abbott DH, Barnett DK, Colman RJ, Yamamoto ME, Schultz-Darken NJ (2003) Aspects of common marmoset basic biology and life history important for biomedical research. Comp Med 53(4):339–350
Tardif SD, Smucny DA, Abbott DH, Mansfield K, Schultz-Darken N, Yamamoto ME (2003) Reproduction in captive common marmosets (Callithrix jacchus). Comp Med 53(4):364–368
Remington ED, Osmanski MS, Wang X (2012) An operant conditioning method for studying auditory behaviors in marmoset monkeys. PLoS One 7(10):e47895
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034
Yperman J, De Visscher G, Holvoet P, Flameng W (2004) Beta-actin cannot be used as a control for gene expression in ovine interstitial cells derived from heart valves. J Heart Valve Dis 13(5):848–853
Ahn K, Huh JW, Park SJ, Kim DS, Ha HS, Kim YJ, Lee JR, Chang KT, Kim HS (2008) Selection of internal reference genes for SYBR green qRT-PCR studies of rhesus monkey (Macaca mulatta) tissues. BMC Mol Biol 9:78
Boda E, Pini A, Hoxha E, Parolisi R, Tempia F (2009) Selection of reference genes for quantitative real-time RT-PCR studies in mouse brain. J Mol Neurosci 37(3):238–253
Brinkhof B, Spee B, Rothuizen J, Penning LC (2006) Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal Biochem 356(1):36–43
Nygard AB, Jorgensen CB, Cirera S, Fredholm M (2007) Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol 8:67
Penna I, Vella S, Gigoni A, Russo C, Cancedda R, Pagano A (2011) Selection of candidate housekeeping genes for normalization in human postmortem brain samples. Int J Mol Sci 12(9):5461–5470
Penning LC, Vrieling HE, Brinkhof B, Riemers FM, Rothuizen J, Rutteman GR, Hazewinkel HA (2007) A validation of 10 feline reference genes for gene expression measurements in snap-frozen tissues. Vet Immunol Immunopathol 120(3–4):212–222
Robinson TL, Sutherland IA, Sutherland J (2007) Validation of candidate bovine reference genes for use with real-time PCR. Vet Immunol Immunopathol 115(1–2):160–165
Bustin S, Penning LC (2012) Improving the analysis of quantitative PCR data in veterinary research. Vet J 191(3):279–281
Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S (2010) MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 11:74
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250
Kessler Y, Helfer-Hungerbuehler AK, Cattori V, Meli ML, Zellweger B, Ossent P, Riond B, Reusch CE, Lutz H, Hofmann-Lehmann R (2009) Quantitative TaqMan real-time PCR assays for gene expression normalisation in feline tissues. BMC Mol Biol 10:106
Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:71
Wood SH, Clements DN, McEwan NA, Nuttall T, Carter SD (2008) Reference genes for canine skin when using quantitative real-time PCR. Vet Immunol Immunopathol 126(3–4):392–395
Suzuki T, Higgins PJ, Crawford DR (2000) Control selection for RNA quantitation. Biotechniques 29:332–337
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75(2–3):291–295
Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M (2000) Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics 2:143–147
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313(4):856–862
Wierschke S, Gigout S, Horn P, Lehmann TN, Dehnicke C, Brauer AU, Deisz RA (2010) Evaluating reference genes to normalize gene expression in human epileptogenic brain tissues. Biochem Biophys Res Commun 403(3–4):385–390
Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GA, Zumla A (2005) The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 344(1):141–143
Aerts JL, Gonzales MI, Topalian SL (2004) Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques 36(1):84–86, 88, 90–81
Al-Bader MD, Al-Sarraf HA (2005) Housekeeping gene expression during fetal brain development in the rat-validation by semi-quantitative RT-PCR. Brain Res Dev Brain Res 156(1):38–45
Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O (2001) Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem 295(1):17–21
Kuchipudi SV, Tellabati M, Nelli RK, White GA, Perez BB, Sebastian S, Slomka MJ, Brookes SM, Brown IH, Dunham SP et al (2012) 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol J 9:230
Schmid H, Cohen CD, Henger A, Irrgang S, Schlondorff D, Kretzler M (2003) Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int 64(1):356–360
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46(1–2):69–81
Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR (2001) Beta-actin–an unsuitable internal control for RT-PCR. Mol Cell Probes 15(5):307–311
Al-Dasooqi N, Bowen JM, Gibson RJ, Logan RM, Stringer AM, Keefe DM (2011) Selection of housekeeping genes for gene expression studies in a rat model of irinotecan-induced mucositis. Chemotherapy 57(1):43–53
Glare EM, Divjak M, Bailey MJ, Walters EH (2002) beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 57(9):765–770
Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, Garrick T, Sheedy D, Hurd Y, Bakalkin G, Ekstrom TJ (2007) Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res 1132(1):20–28
Acknowledgments
This work was supported partly by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to Y. S. (no. 23658255).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shimamoto, Y., Kitamura, H., Niimi, K. et al. Selection of suitable reference genes for mRNA quantification studies using common marmoset tissues. Mol Biol Rep 40, 6747–6755 (2013). https://doi.org/10.1007/s11033-013-2791-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2791-0