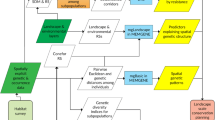
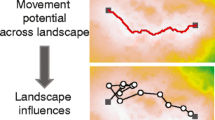
Abstract
Understanding how landscape heterogeneity constrains gene flow and the spread of adaptive genetic variation is important for biological conservation given current global change. However, the integration of population genetics, landscape ecology and spatial statistics remains an interdisciplinary challenge at the levels of concepts and methods. We present a conceptual framework to relate the spatial distribution of genetic variation to the processes of gene flow and adaptation as regulated by spatial heterogeneity of the environment, while explicitly considering the spatial and temporal dynamics of landscapes, organisms and their genes. When selecting the appropriate analytical methods, it is necessary to consider the effects of multiple processes and the nature of population genetic data. Our framework relates key landscape genetics questions to four levels of analysis: (i) node-based methods, which model the spatial distribution of alleles at sampling locations (nodes) from local site characteristics; these methods are suitable for modeling adaptive genetic variation while accounting for the presence of spatial autocorrelation. (ii) Link-based methods, which model the probability of gene flow between two patches (link) and relate neutral molecular marker data to landscape heterogeneity; these methods are suitable for modeling neutral genetic variation but are subject to inferential problems, which may be alleviated by reducing links based on a network model of the population. (iii) Neighborhood-based methods, which model the connectivity of a focal patch with all other patches in its local neighborhood; these methods provide a link to metapopulation theory and landscape connectivity modeling and may allow the integration of node- and link-based information, but applications in landscape genetics are still limited. (iv) Boundary-based methods, which delineate genetically homogeneous populations and infer the location of genetic boundaries; these methods are suitable for testing for barrier effects of landscape features in a hypothesis-testing framework. We conclude that the power to detect the effect of landscape heterogeneity on the spatial distribution of genetic variation can be increased by explicit consideration of underlying assumptions and choice of an appropriate analytical approach depending on the research question.


Similar content being viewed by others
References
Anderson CD, Epperson BK, Fortin MJ, Holderegger R, James PMA, Rosenber MS, Sribner KT, Spear SF (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol 19:3565–3575
Angelone S, Kienast F, Holderegger R (2011) Where movement happens: scale-dependent landscape effects on genetic differentiation in the European tree frog. Ecography 34:714–722
Balkenhol N, Gugerli F, Cushman SA, Waits L, Coulon A, Arntzen J, Holderegger R, Wagner HH (2009a) Identifying future research needs in landscape genetics: where to from here? Landsc Ecol 24:455–463
Balkenhol N, Waits LP, Dezzani RJ (2009b) Statistical approaches in landscape genetics: an evaluation of methods for linking landscape and genetic data. Ecography 32:818–830
Barbujani G, Sokal RR (1991) Genetic population structure of Italy. II. Physical and cultural barriers to gene flow. Am J Hum Genet 48:398–411
Barbujani G, Oden NL, Sokal RR (1989) Detecting regions of abrupt change in maps of biological variables. Syst Zool 38:376–389
Borcard D, Gillet F, Legendre P (2011) Numerical ecology with R. Springer, New York
Bulman CR, Wilson RJ, Holt AR, Bravo LG, Early RI, Warren MS, Thomas CD (2007) Minimum viable metapopulation size, extinction debt, and the conservation of a declining species. Ecol Appl 17:1460–1473
Cushman SA, Landguth EL (2010) Spurious correlations and inference in landscape genetics. Mol Ecol 19:3592–3602
Cushman S, McKelvey K, Hayden J, Schwartz M (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Dale MRT, Fortin MJ (2010) From graphs to spatial graphs. Annu Rev Ecol Evol Syst 41:21–38
DalGrande F, Widmer I, Wagner HH, Scheidegger C (2012) Vertical and horizontal photobiont transmission within populations of a lichen symbiosis. Mol Ecol. doi:10.1111/j.1365-294X.2012.05482.x
Dawson KJ, Belkhir K (2001) A Bayesian approach to the identification of panmictic populations and the assignment of individuals. Genet Res 78:59–77
Diniz-Filho JAF, Nabout JC, Telles MPdC, Soares TN, Rangel TFLVB (2009) A review of techniques for spatial modeling in geographical, conservation and landscape genetics. Genet Mol Biol 32:203–211
Dray S, Pélissier R, Couteron P, Fortin MJ, Legendre P, Peres-Neto PR, Bellier E, Bivand R, Blanchet FG, De Cáceres M, Dufour A, Heegard E, Jombart T, Munoz F, Oksanen J, Thioulouse J, Wagner HH. Community ecology in the age of multivariate multiscale spatial analysis. Ecol Monogr. doi:10.1890/11-1183.1
Dray S, Legendre P, Peres-Neto PR (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196:483–493
Durand E, Jay F, Gaggiotti OE, François O (2009) Spatial inference of admixture proportions and secondary contact zones. Mol Biol Evol 26:1963–1973
Dyer RJ, Nason JD, Garrick RC (2010) Landscape modelling of gene flow: improved power using conditional genetic distance derived from the topology of population networks. Mol Ecol 19:3746–3759
Epperson BK (2003) Geographical genetics. Princeton University Press, Princeton
Folke C, Carpenter S, Walker B, Scheffer M, Elmqvist T, Gunderson L, Holling CS (2004) Regime shifts, resilience, and biodiversity in ecosystem management. Annu Rev Ecol Evol Syst 35:557–581
Foll M, Gaggiotti OE (2006) Identifying the environmental factors that determine the genetic structure of populations. Genetics 174:875–891
Fortin MJ, Dale MRT (2005) Spatial analysis: a guide for ecologists. Cambridge University Press, New York
Fortin MJ, Drapeau P, Jacquez GM (1996) Quantification of the spatial co-occurrences of ecological boundaries. Oikos 77:51–60
François O, Durand E (2010) Spatially explicit Bayesian clustering models in population genetics. Mol Ecol Resour 10:773–784
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Frankham R (2009) Introduction to conservation genetics. Cambridge University Press, New York
Garant D, Kruuk LEB, McCleery RH, Sheldon BC (2007) The effects of environmental heterogeneity on multivariate selection on reproductive traits in female great tits. Evolution 61:1546–1559
Garroway CJ, Bowman J, Wilson PJ (2011) Using a genetic network to parameterize a landscape resistance surface for fishers, Martes pennanti. Mol Ecol 20:3978–3988
Greenwald KR (2010) Genetic data in population viability analysis: case studies with ambystomatid salamanders. Anim Conserv 13:115–122
Holderegger R, Wagner HH (2008) Landscape genetics. Bioscience 58:199–207
Holderegger R, Buehler D, Gugerli F, Manel S (2010) Landscape genetics of plants. Trends Plant Sci 15:675–683
Holling CS (1992) Cross-scale morphology, geometry, and dynamics of ecosystems. Ecol Monogr 62:447–502
Jacquez GM, Maruca S, Fortin MJ (2000) From fields to objects: a review of geographic boundary analysis. J Geogr Syst 2:221–241
James PMA, Coltman DW, Murray BW, Hamelin RC, Sperling FAH (2011) Spatial genetic structure of a symbiotic beetle-fungal system: toward multi-taxa integrated landscape genetics. PLoS One 6:e25359
Jay F (2011) PoPS: prediction of population genetic structure—program documentation and tutorial. University Joseph Fourier, Grenoble
Keyghobadi N, Roland J, Matter SF, Strobeck C (2005) Among- and within-patch components of genetic diversity respond at different rates to habitat fragmentation: an empirical demonstration. Proc Biol Sci 272:553–560
Lande R (1991) Isolation by distance in a quantitative trait. Genetics 128:443–452
Landguth EL, Cushman S, Murphy MA, Luikart G (2010) Relationships between migration rates and landscape resistance assessed using individual-based simulations. Mol Ecol Resour 10:854–862
Legendre P, Fortin MJ (2010) Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol Ecol Resour 10:831–844
Legendre P, Legendre L (1998) Numerical ecology. Elsevier, New York
Lichstein JW, Simons TR, Shriner SA, Franzreb KE (2002) Spatial autocorrelation and autoregressive models in ecology. Ecol Monogr 72:445–463
Manel S, Joost S, Epperson BK, Holderegger R, Storfer A, Rosenberg MS, Scribner KT, Bonin A, Fortin MJ (2010) Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol Ecol 19:3760–3772
McRae BH, Dickson BG, Keitt TH, Shah VB (2008) Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89:2712–2724
Méndez M, Tella JL, Godoy JA (2011) Restricted gene flow and genetic drift in recently fragmented populations of an endangered steppe bird. Biol Conserv 144:2615–2622
Monmonier MS (1973) Maximum-difference barriers: an alternative numerical regionalization method. Geogr Anal 5:245–261
Murphy MA, Dezzani RJ, Pilliod DS, Storfer A (2010) Landscape genetics of high mountain frog metapopulations. Mol Ecol 19:3634–3649
Oden NL, Sokal R, Fortin MJ, Goebl H (1993) Categorical wombling: detecting regions of significant change in spatially located categorical variables. Geogr Anal 25:315–336
Safner T, Miller MP, McRae BH, Fortin MJ, Manel S (2011) Comparison of bayesian clustering and edge detection methods for inferring boundaries in landscape genetics. Int J Mol Sci 12:865–889
Slatkin M, Arter HE (1991) Spatial autocorrelation methods in population genetics. Am Nat 138:499–517
Sokal RR (1979) Testing statistical significance of geographic variation patterns. Syst Zool 28:227–232
Spear SF, Balkenhol N, Fortin MJ, McRae BH, Scribner KT (2010) Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol Ecol 19:3576–3591
Spielman D, Brook BW, Frankham R, Schaal BA (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101:15261–15264
St-Louis V, Fortin MJ, Desrochers A (2004) Spatial association between forest heterogeneity and breeding territory boundaries of two forest songbirds. Landsc Ecol 19:591–601
Storfer A, Murphy MA, Evans JS, Goldberg CS, Robinson S, Spear SF, Dezzani R, Delmelle E, Vierling L, Waits LP (2007) Putting the ‘landscape’ in landscape genetics. Heredity 98:128–142
Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP (2010) Landscape genetics: where are we now? Mol Ecol 19:3496–3514
Wagner HH, Fortin MJ (2005) Spatial analysis of landscapes: concepts and statistics. Ecology 86:1975–1987
Ward EJ (2008) A review and comparison of four commonly used Bayesian and maximum likelihood model selection tools. Ecol Model 211:1–10
Wright S (1943) Isolation by distance. Genetics 28:114–138
Wright S (1948) On the roles of directed and random changes in gene frequency in the genetics of populations. Evolution 2:279–294
Acknowledgments
This work resulted from a Distributed Graduate Seminar (Developing Best Practices for Testing Landscape Effects on Gene Flow), conducted through the National Center for Ecological Analysis and Synthesis, a center funded by the National Science Foundation grant #EF-0553768, the University of California, Santa Barbara, and the State of California. It was also supported by NSERC discovery grants to HHW and MJF. We thank Michelle DiLeo and Yessica Rico for their thoughtful comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wagner, H.H., Fortin, MJ. A conceptual framework for the spatial analysis of landscape genetic data. Conserv Genet 14, 253–261 (2013). https://doi.org/10.1007/s10592-012-0391-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0391-5