Abstract
Clinical guidelines for breast cancer treatment differ in their selection of patients at a high risk of recurrence who are eligible to receive adjuvant systemic treatment (AST). The 70-gene signature is a molecular tool to better guide AST decisions. The aim of this study was to evaluate whether adding the 70-gene signature to clinical risk prediction algorithms can optimize outcome prediction and consequently treatment decisions in early stage, node-negative breast cancer patients. A 70-gene signature was available for 427 patients participating in the RASTER study (cT1-3N0M0). Median follow-up was 61.6 months. Based on 5-year distant-recurrence free interval (DRFI) probabilities survival areas under the curve (AUC) were calculated and compared for risk estimations based on the six clinical risk prediction algorithms: Adjuvant! Online (AOL), Nottingham Prognostic Index (NPI), St. Gallen (2003), the Dutch National guidelines (CBO 2004 and NABON 2012), and PREDICT plus. Also, survival AUC were calculated after adding the 70-gene signature to these clinical risk estimations. Systemically untreated patients with a high clinical risk estimation but a low risk 70-gene signature had an excellent 5-year DRFI varying between 97.1 and 100 %, depending on the clinical risk prediction algorithms used in the comparison. The best risk estimation was obtained in this cohort by adding the 70-gene signature to CBO 2012 (AUC: 0.644) and PREDICT (AUC: 0.662). Clinical risk estimations by all clinical algorithms improved by adding the 70-gene signature. Patients with a low risk 70-gene signature have an excellent survival, independent of their clinical risk estimation. Adding the 70-gene signature to clinical risk prediction algorithms improves risk estimations and therefore might improve the identification of early stage node-negative breast cancer patients for whom AST has limited value. In this cohort, the PREDICT plus tool in combination with the 70-gene signature provided the best risk prediction.
Similar content being viewed by others
Introduction
For the past decade, the selection of early stage breast cancer patients who are at a high risk of recurrence and eligible to receive adjuvant systemic treatment (AST) is based on clinicopathological factors, such as age, tumor size, nodal status, histological grade, and hormone-receptor status. Several clinical risk prediction algorithms used in online tools and guidelines, such as Adjuvant! Online (AOL), the Nottingham Prognostic Index (NPI), the St. Gallen expert panel recommendations of 2003, and the Dutch national guidelines of 2004 and 2012, use these factors in specific algorithms for risk estimations and AST recommendations [1–6]. A relatively new online tool for outcome prediction in breast cancer patients is PREDICT plus [7]. This tool not only uses the clinicopathological factors mentioned above, but also incorporates human epidermal growth factor receptor 2 (HER2) status and method of detection. Both of these factors have proven to be independent prognostic factors in overall and breast cancer-specific survival [7, 8].
Even with the aid of these clinical risk prediction algorithms, individual risk assessment remains challenging. Each of these clinical risk prediction algorithms may define a slightly different group of patients at a low or high risk, which are partly non-overlapping. This indicates that it is unclear which tool or guideline has the highest prognostic accuracy for the individual patient [1, 5, 6, 9]. Moreover, online tools such as AOL provide a survival probability without stratification into high versus low risk. The choice for a specific cut-off point in risk clearly influences the concordance with other tools [10]. Gene-expression classifiers have been developed and validated on historic data to refine clinical risk estimations and related AST recommendations [11, 12]. One of these classifiers is the 70-gene signature (MammaPrint™, Agendia Inc., Amsterdam, the Netherlands) [13, 14]. Between 2004 and 2006, the 70-gene signature has been assessed in the first prospective study using a gene-expression classifier as a risk estimation tool in addition to clinicopathological factors to determine the need for AST. A considerable discrepancy in risk estimations among different clinical guidelines and the 70-gene signature was observed [9, 15]. Recently, the 5-year follow-up data of the RASTER study were reported showing an excellent distant-recurrence free interval (DRFI) of 97 % for patients with a low risk 70-gene signature. Patients with a high risk 70-gene signature showed a DRFI of 92 % [16]. When compared to AOL, 70-gene signature low-AOL high risk patients who did not receive any AST showed a DRFI of 100 %. This indicates that omission of chemotherapy in these patients may not compromise outcome. Up to the evaluated 5-year median survival, the number of events is small and the follow-up time is relatively short. However, AOL is not the only risk estimation tool used in clinical practice today. In addition, the 70-gene signature is more likely to be added to clinical risk prediction algorithms instead of replacing them. Therefore, we evaluated whether adding the 70-gene signature to clinical risk prediction algorithms can improve individual outcome prediction in early stage, node-negative breast cancer patients.
Patients and methods
The RASTER study design, patient eligibility criteria, and study logistics have been described elsewhere (www.controlled-trials.com/ISRCTN71917916) [15]. In short, 812 female patients were enrolled in 16 hospitals in the Netherlands. 427 patients were postoperatively eligible and for them a 70-gene signature (MammaPrint™, Agendia Inc.) was obtained. All patients were between 18 and 61 years old and had a histologically confirmed unilateral, unifocal, primary operable, invasive adenocarcinoma of the breast (cT1-3N0M0). All patients were primarily surgically treated with either breast-conserving surgery or mastectomy. To insure routine clinical practice, the initial histopathology data were used for clinical risk assessment by the treating physician and in the statistical analysis, without central review of the paraffin-embedded tumor samples. Details on tumor grading, assessment of hormone-receptor status and HER2 status, RNA extraction and microarray analysis have been described elsewhere [15]. Decisions on whether or not to treat with AST (comprising chemotherapy and/or endocrine therapy) in the RASTER study were based on the Dutch national guidelines of 2004, the 70-gene signature, and doctors’ and patients’ preferences [15]. More detailed insight on the follow-up data of this cohort is described elsewhere [16].
Clinical risk prediction algorithms
Hereafter, risk assessment by use of clinicopathological factors is referred to as “clinical risk.” Guidelines used in this study to assess clinical risk were Adjuvant! Online (AOL), Nottingham Prognostic Index (NPI), the St. Gallen expert panel recommendations (2003, current at the time the RASTER study was conducted), the Dutch national guidelines (2004, current at the time the RASTER study was conducted, and 2012), and PREDICT plus. Adjuvant! Online software, version 8.0, calculates the 10-year survival probabilities based on the age of the patient, tumor size, tumor grade, estrogen receptor (ER) status, and nodal status [5, 10]. Patients were considered high risk if their calculated 10-year survival probability was less than 90 % [15]. This cut off was also used in the RASTER study and similar to the cut off used in the MINDACT trial. The NPI computes a score with the algorithm: 0.2 × size (cm) + grade + nodal status. A moderate or high risk was defined as a score greater than 3.4 [1, 17]. The St. Gallen expert panel of 2003 recommended to define low clinical risk as ER positive or progesterone receptor (PR)-positive status (or both) and all the following criteria: tumor size of 2 cm or smaller, grade 1, and age 35 years or over. All other tumors were deemed to be associated with a moderate or high risk of distant metastasis and death [2]. The 2004 Dutch national guidelines define high clinical risk for node-negative breast cancer as age 35 years or younger (except for tumors grade 1 of 10 mm or smaller), a tumor of grade 3 and 10 mm or larger, or grade 2 and 20 mm or larger, and every tumor larger than 30 mm. Adjuvant endocrine treatment was advised only in clinically high risk patients with hormone-receptor-positive tumors in combination with chemotherapy [10]. AST was justified for patients with a 10-year survival probability of less than 80 %. The less restrictive Dutch guidelines of 2012 define high clinical risk for node-negative breast cancer as age under 35 years except for tumors grade 1 of 10 mm or smaller, or age 35 years or older with a tumor of grade 2 or higher, and 10–20 mm in size, and every tumor larger than 20 mm. According to this 2012 guideline, AST was justified for patients with a 10-year survival probability of less than 85 %. The online PREDICT plus tool estimates the 5- and 10-year survival probabilities based on the age of the patient, method of detection, tumor size, tumor grade, number of positive nodes, ER and HER2 status [7]. We defined a 5-year survival probability of <95 %, which is in line with the cut offs used for Adjuvant! Online. All clinicopathological factors used by the guidelines mentioned above were summarized elsewhere [18]. In our analyses, a moderate or high clinical risk was considered an indication for AST.
Statistical analysis
We estimated a 5-year DRFI, comprising distant recurrence and death from breast cancer [19]. Survival curves were constructed using the Kaplan–Meier method and compared using the log-rank test. Survival ROC and AUC (c-index) analyses were performed to evaluate the additional value of the 70-gene signature to the clinical guidelines described in this manuscript. An ANOVA test was used to compare the model before and after adding the 70-gene signature. A significant finding was defined as a p value below 0.05. Analyses were performed using SAS version 9.2 and R version 2.14.0.
Results
Patient and tumor characteristics, AST and outcome stratified by 70-gene signature
Patient and tumor characteristics were described elsewhere [15]. After a median follow-up time of 61.6 months, 24 DRFI events occurred. Eleven patients died of whom nine due to breast cancer. The 5-year DRFI probabilities for 70-gene signature low risk (n = 219) and high risk (n = 208) patients were 97.0 % (95 % CI 94.7–99.4) and 91.7 % (95 % CI 87.9–95.7) (p = 0.03), respectively (Supplementary Fig. 1) [16].
Additional value of 70-gene signature to clinical risk assessment
Adding the 70-gene signature to clinical risk prediction algorithms improved outcome prediction. For most guidelines, this was a borderline significant improvement of the c-index (Table 1). The c-index was highest for PREDICT plus (0.627), followed by NPI (0.591), and the Dutch national guidelines of 2004 (0.586). Adding the 70-gene signature improved the model to 0.638 for NPI (p = 0.05) and to 0.639 for the Dutch national guidelines of 2004 (p = 0.04). The best risk predictions were achieved when using PREDICT plus (0.662) or the Dutch guidelines of 2012 (0.644) in combination with the 70-gene signature. The c-index for AOL was lowest, before (0.532) and after adding the 70-gene signature (0.619).
Discordance between clinical risk assessment and the 70-gene signature
Discordant risk estimations occurred in 37 % of the cases (161/427) for AOL, 27 % for NPI (117/427), 39 % for St. Gallen (168/427), 30 % for the Dutch national guidelines of 2004 (128/427), 39 % for the guidelines of 2012 (167/427), and 25 % for PREDICT plus (107/427) (Table 2; Fig. 1). Most discordant cases were 70-gene signature low risk and clinically high risk; 29 % for AOL (124/427), 10 % for NPI (44/427), 37 % for St. Gallen (157/427), 12 % for the Dutch national guidelines of 2004 (52/427), 31 % for the guidelines of 2012 (131/427), and 11 % for PREDICT plus at 5 years (49/427).
Table 2 summarizes the AST given in the different categories stratified by 70-gene signature and clinical risk. When the 70-gene signature was used, 20 % less patients would be eligible to receive ACT compared to AOL, 34 % less compared to St. Gallen, 6 % less compared to the Dutch guidelines of 2004, and 22 % less compared to the guidelines of 2012. The 70-gene signature identifies 7 % more patients eligible to receive ACT compared to NPI and 2 % more compared to PREDICT plus.
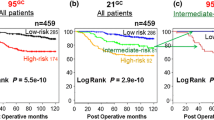
The 5-year DRFI probabilities for AOL low risk (n = 132) and high risk (n = 295) patients were 96.7 % (95 % CI 93.5–100) and 93.4 % (95 % CI 90.4–96.4), respectively (p = 0.24). For NPI low risk (n = 248) and high risk (n = 179) patients, the 5-year DRFI probabilities were 96.7 % (95 % CI 94.2–99.2) and 91.3 % (95 % CI 87.2–95.6) (p = 0.03). The St. Gallen low risk (n = 73) and high risk (n = 353) patients showed 5-year DRFI probabilities of 98.5 % (95 % CI 95.7–100) and 93.5 % (95 % CI 90.9–96.3) (p = 0.08). For the Dutch national guidelines of 2004 low risk (n = 243) and high risk (n = 184) patients, the 5-year DRFI probabilities were 96.6 % (95 % CI 94.2–99.2) and 91.5 % (95 % CI 87.4–95.7), respectively (p = 0.11), while for the Dutch national guidelines of 2012 low risk (n = 124) and high risk (n = 303) patients the 5-year DRFI probabilities were 99.2 % (95 % CI 97.6–100) and 92.4 % (95 % CI 89.3–95.6) (p = 0.02). The 5-year prediction of PREDICT plus low risk (n = 228) and high risk (n = 199) patients showed 5-year DRFI probabilities of 96.8 % (95 % CI 94.2–99.4) and 91.7 % (95 % CI 87.9–95.7), respectively (p = 0.004) (Fig. 2). Table 3 summarizes DRFI probabilities according to the combined risk categories.
Subgroup analyses of therapy-naïve patients
Of the patients who had a low risk 70-gene signature 85 % did not receive adjuvant chemotherapy. Only 27 % of the 70-gene signature low risk patients received adjuvant endocrine therapy. Among the low risk systemically untreated patients, no significant difference was seen for most clinical risk algorithms (p = 0.29 for AOL, p = 0.66 for NPI, p = 0.37 for St. Gallen, p = 0.65 for the 2004, and p = 0.14 for the 2012 Dutch national guidelines) between patients with a concordant low risk assessment and patients with a 70-gene signature low risk result but a high risk assessment by one or more of the clinical indexes (Fig. 1). Only the PREDICT plus tool shows that patients with a concordant low risk assessment (n = 141) at 5 years have a significantly better DRFI survival probability compared to patients with a low risk 70-gene signature and a high risk according to PREDICT plus (n = 17) (p = 0.002).
Discussion
The RASTER study was the first study to prospectively evaluate the outcome of patients for whom the 70-gene signature was used for risk estimations and AST recommendations. The recently published 5-year follow-up data of this study provide the opportunity to evaluate the additional value of a gene-expression classifier to risk estimations based on clinicopathological factors incorporated in clinical tools and guidelines. Of all clinical risk prediction algorithms used in this study, the online PREDICT plus tool provided the best risk estimation. Addition of the 70-gene signature to either the PREDICT plus tool or the Dutch national guidelines of 2012 resulted in the best risk estimations in this cohort. Interestingly, AOL showed the lowest c-index before and after adding the 70-gene signature. This might be explained by the fact that this guideline does not incorporate HER2 status, while the Dutch guidelines of 2012 and PREDICT plus do take this clinicopathological factor into account. In addition, as AOL does not provide a classification into high versus low risk, the choice for a specific cut-off point may influence these results. Previous analyses already showed that method of detection is an independent prognostic factor in breast cancer-specific and overall survival. The fact that the PREDICT plus tool takes the method of detection into account may explain why this risk prediction algorithm performs so well in this cohort. When solely using the 70-gene signature, the number of patients at high risk of recurrence who are eligible for adjuvant chemotherapy would be reduced by 20 % compared to AOL. As a similar comparison was made in the MINDACT trial (AOL in MINDACT does include HER2), one can hypothesize that a similar reduction in chemotherapy will be seen in this large, randomized controlled phase 3 trial. Analyses of the first 800 patients included in the MINDACT trial show a similar possible reduction in adjuvant chemotherapy of 18 % (141/800). Overall, the 5-year outcome of this cohort of patients for whom the 70-gene signature result was prospectively used to guide AST decisions was favorable. One should take into consideration that a substantial proportion of patients, 39 % (168/427) of this cohort, did not receive any form of AST. Most importantly, the 5-year DRFI probabilities were excellent for patients who were clinically at high risk but had a low risk 70-gene signature, even in the absence of any AST [16]. Therefore, omission of chemotherapy in patients with a low risk 70-gene signature appeared safe, even in case of a high risk estimation by one or more of the clinical guidelines. A larger number of patients in the untreated subgroups and longer follow-up are needed to draw firm conclusions. The only tool that was able to select patients at a slightly higher risk of recurrence among the 70-gene signature low risk patients was the PREDICT plus tool. However, in this subgroup the number of patients (n = 17) was too low to draw any firm conclusions. A larger cohort is necessary to evaluate the additional prognostic value of the 70-gene signature to PREDICT plus tool. An advantage, but also a limitation of this study is that the actual treatment decisions were based on the Dutch guidelines of 2004, the 70-gene signature result and preferences of doctors and patients. The study design provides an optimal reflection of daily clinical practice, but subtle selection mechanisms may be present and may have influenced our results. Another possible limitation is that all clinical tools and guidelines included in our analyses use slightly different definitions of high and low risk. These differences create an additional group of patients for whom the guidelines provide discordant risk estimations. Also, some guidelines base their risk assessment on 5-year survival probabilities, while others on 10-year survival probabilities. In our analyses, we were unable to adjust for these differences which make a head-to-head comparison more difficult to interpret. Still, the guidelines as used in this study reflect the way they are used in current daily clinical practice. The c-indexes reported here leave room for improvement and this again underlines the need for more accurate, personalized breast cancer care. Also, it should be kept in mind that the results of this study are based on a case mix of relatively young (<61 years) breast cancer patients. Finally, central pathology revision might have changed the results, since an earlier report showed that for 8 % of the patients AOL risk estimations would change based on revised pathology [20].
In conclusion, our results indicate that adding the 70-gene signature clinical guidelines with the 70-gene signature improves risk estimations and therefore may help to identify early stage node-negative breast cancer patients for whom limited AST might be appropriate and for whom overtreatment can be avoided. In this cohort, PREDICT plus appeared to be a promising tool to identify patients for whom limited AST in case of early stage node-negative disease might be appropriate.
References
D’Eredita’ G, Giardina C, Martellotta M, Natale T, Ferrarese F (2001) Prognostic factors in breast cancer: the predictive value of the Nottingham Prognostic Index in patients with a long-term follow-up that were treated in a single institution. Eur J Cancer 37:591–596
Goldhirsch A, Glick JH, Gelber RD, Coates AS, Senn HJ (2001) Meeting highlights: International Consensus Panel on the treatment of primary breast cancer. Seventh international conference on adjuvant therapy of primary breast cancer. J Clin Oncol 19:3817–3827
Integraal Kankercentrum Nederland: NABON richtlijn mammacarcinoom (2012)
Kwaliteitsinstituut voor de Gezondheidszorg CBO VvlK: Adjuvante Systemische Therapie voor het Operabel Mammacarcinoom (2004) Richtlijn Behandeling van het Mammacarcinoom 46–70
Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, Norris BD, Davis GJ, Chia SK, Gelmon KA (2005) Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol 23:2716–2725
Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thurlimann B, Senn HJ (2003) Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. J Clin Oncol 21:3357–3365
Wishart GC, Bajdik CD, Dicks E, Provenzano E, Schmidt MK, Sherman M, Greenberg DC, Green AR, Gelmon KA, Kosma VM, Olson JE, Beckmann MW, Winqvist R, Cross SS, Severi G, Huntsman D, Pylkas K, Ellis I, Nielsen TO, Giles G, Blomqvist C, Fasching PA, Couch FJ, Rakha E, Foulkes WD, Blows FM, Begin LR, van’t Veer LJ, Southey M, Nevanlinna H, Mannermaa A, Cox A, Cheang M, Baglietto L, Caldas C, Garcia-Closas M, Pharoah PD (2012) PREDICT plus: development and validation of a prognostic model for early breast cancer that includes HER2. Br J Cancer 107:800–807
Mook S, van’t Veer LJ, Rutgers EJ, Ravdin PM, van de Velde AO, van Leeuwen FE, Visser O, Schmidt MK (2011) Independent prognostic value of screen detection in invasive breast cancer. J Natl Cancer Inst 103:585–597
Bueno-de-Mesquita JM, Sonke GS, van de Vijver MJ, Linn SC (2011) Additional value and potential use of the 70-gene prognosis signature in node-negative breast cancer in daily clinical practice. Ann Oncol 22:2021–2030
Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, Parker HL (2001) Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol 19:980–991
Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris A, Bogaerts J, Therasse P, Floore A, Amakrane M, Piette F, Rutgers E, Sotiriou C, Cardoso F, Piccart MJ (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 98:1183–1192
Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN (2008) Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist 13:477–493
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009
Bueno-de-Mesquita JM, van Harten WH, Retel VP, van’t Veer LJ, van Dam FS, Karsenberg K, Douma KF, van Tinteren H, Peterse JL, Wesseling J, Wu TS, Atsma D, Rutgers EJ, Brink G, Floore AN, Glas AM, Roumen RM, Bellot FE, van Krimpen C, Rodenhuis S, van de Vijver MJ, Linn SC (2007) Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: a prospective community-based feasibility study (RASTER). Lancet Oncol 8:1079–1087
Drukker CA, Bueno-de-Mesquita JM, Retel VP, van Harten WH, van Tinteren H, Wesseling J, Roumen RM, Knauer M, van’t Veer LJ, Sonke GS, Rutgers EJ, van de Vijver MJ, Linn SC (2013) A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer 133:929–936
Todd JH, Dowle C, Williams MR, Elston CW, Ellis IO, Hinton CP, Blamey RW, Haybittle JL (1987) Confirmation of a prognostic index in primary breast cancer. Br J Cancer 56:489–492
Drukker CA, van den Hout HC, Sonke GS, Brain E, Bonnefoi H, Cardoso F, Goldhirsch A, Harbeck N, Honkoop AH, Koornstra RH, van Laarhoven HWM, Portielje JEA, Schneeweiss A, Smorenburg CH, Stouthard J, Linn SC, Schmidt MK (2014) Risk estimations and treatment decisions in early stage breast cancer; agreement among oncologists and the impact of the 70-gene signature. Eur J Cancer 50(6):1045–1054
Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, Sparano JA, Hunsberger S, Enos RA, Gelber RD, Zujewski JA (2007) Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol 25:2127–2132
Bueno-de-Mesquita JM, Nuyten DS, Wesseling J, van Tinteren H, Linn SC, van de Vijver MJ (2010) The impact of inter-observer variation in pathological assessment of node-negative breast cancer on clinical risk assessment and patient selection for adjuvant systemic treatment. Ann Oncol 21:40–47
Acknowledgments
We are indebted to the women who participated in the RASTER study; to the doctors, nurses, and data managers from the participating hospitals in the Netherlands that enrolled patients in the RASTER-study and contributed to the collection of follow-up data.
Conflict of interest
The RASTER study was financially supported by the Dutch Health Care Insurance Board (CVZ). LJvV and MJvdV are named inventors on the patent for the 70-gene signature used in this study. LJvV reports being shareholder in and part-time employed by Agendia Inc., the commercial company that markets the 70-gene signature as MammaPrint™. LJvV was supported by the Dutch Genomics Initiative “Cancer Genomics Centre.”
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Drukker, C.A., Nijenhuis, M.V., Bueno-de-Mesquita, J.M. et al. Optimized outcome prediction in breast cancer by combining the 70-gene signature with clinical risk prediction algorithms. Breast Cancer Res Treat 145, 697–705 (2014). https://doi.org/10.1007/s10549-014-2954-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-2954-2