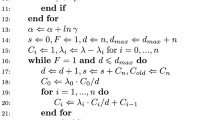Abstract
Data augmentation (DA) is a flexible tool for analyzing closed and open population models of capture–recapture data, especially models which include sources of hetereogeneity among individuals. The essential concept underlying DA, as we use the term, is based on adding “observations” to create a dataset composed of a known number of individuals. This new (augmented) dataset, which includes the unknown number of individuals N in the population, is then analyzed using a new model that includes a reformulation of the parameter N in the conventional model of the observed (unaugmented) data. In the context of capture–recapture models, we add a set of “all zero” encounter histories which are not, in practice, observable. The model of the augmented dataset is a zero-inflated version of either a binomial or a multinomial base model. Thus, our use of DA provides a general approach for analyzing both closed and open population models of all types. In doing so, this approach provides a unified framework for the analysis of a huge range of models that are treated as unrelated “black boxes” and named procedures in the classical literature. As a practical matter, analysis of the augmented dataset by MCMC is greatly simplified compared to other methods that require specialized algorithms. For example, complex capture–recapture models of an augmented dataset can be fitted with popular MCMC software packages (WinBUGS or JAGS) by providing a concise statement of the model’s assumptions that usually involves only a few lines of pseudocode. In this paper, we review the basic technical concepts of data augmentation, and we provide examples of analyses of closed-population models (M 0, M h, distance sampling, and spatial capture–recapture models) and open-population models (Jolly–Seber) with individual effects.



Similar content being viewed by others
References
Bled F, Royle JA, Cam E (2010) Hierarchical modeling of an invasive spread: case of the Eurasian collared-dove Streptopelia decaocto in the USA. Ecol Appl (in press)
Bonner S, Schwarz C (2006) An extension of the Cormack Jolly Seber model for continuous covariates with application to Microtus pennsylvanicus. Biometrics 62:142–149
Borchers DL, Efford MG (2008) Spatially explicit maximum likelihood methods for capture–recapture studies. Biometrics 64:377–385
Burnham KP, Overton WS (1978) Estimation of the size of a closed population when capture probabilities vary among animals. Biometrika 65:625–633
Converse SJ, Royle JA (2010) Dealing with incomplete and variable detectability in multi-year, multi-site monitoring of ecological populations. In: Design and analysis of long-term ecological monitoring studies (in press)
Cooch E, White G (2001) Using MARK: a gentle introduction. Cornell University, Ithaca
Coull BA, Agresti A (1999) The use of mixed logit models to reflect heterogeneity in capture–recapture studies. Biometrics 55:294–301
Crosbie SF, Manly BFJ (1985) Parsimonious modelling of capture–mark-recapture studies. Biometrics 41:385–398
Dorazio RM, Royle JA (2003) Mixture models for estimating the size of a closed population when capture rates vary among individuals. Biometrics 59:350–363
Dorazio RM, Royle JA (2005) Estimating size and composition of biological communities by modeling the occurrence of species. J Am Stat Assoc 100:389–398
Dorazio RM, Royle JA, Soderstrom B, Glimskar A (2006) Estimating species richness and accumulation by modeling species occurrence and detectability. Ecology 87:842–854
Dorazio RM, Kéry M, Royle JA, Plattner M (2010) Models for inference in dynamic metacommunity systems. Ecology 91:2466–2475
Dupuis JA, Schwarz CJ (2007) A Bayesian approach to the multistate Jolly-Seber capture–recapture model. Biometrics 63:1015–1022
Durban JW, Elston DA (2005) Mark-recapture with occasion and individual effects: abundance estimation through Bayesian model selection in a fixed dimensional parameter space. J Agric Biol Environ Stat 10:291–305
Efford M (2004) Density estimation in live-trapping studies. Oikos 106:598–610
Gardner B, Royle JA, Wegan MT, Rainbolt RE, Curtis PD (2010) Estimating black bear density using DNA data from hair snares. J Wildl Manag 74:318–325
Gardner B, Reppucci J, Lucherini M, Royle JA (2010b) Spatially-explicit inference for open populations: estimating demographic parameters from camera-trap studies. Ecology 91:3376–3383
Gimenez O, Rossi V, Choquet R, Dehais C, Doris B, Varella H, Vila JP, Pradel R (2007) State-space modelling of data on marked individuals. Ecol Model 206:431–438
Gimenez O, Choquet R (2010) Individual heterogeneity in studies on marked animals using numerical integration: capture–recapture mixed models. Ecology 91:148–154
Green PJ (1995) Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82:711–732
Johnson DH (1999) The insignificance of statistical significance testing. J Wildl Manag 63:763–772
Jolly G (1965) Explicit estimates from capture–recapture data with both death and immigration—stochastic model. Biometrika 52:225–247
Karanth KU (1995) Estimating tiger (Panthera tigris) populations from camera-trap data using capture–recapture models. Biol Conserv 71:333–338
Karanth KU, Nichols JD (1998) Estimation of tiger densities in India using photographic captures and recaptures. Ecology 79:2852–2862
Karanth K, Nichols JD, Kumar N, Hines JE (2006) Assessing tiger population dynamics using photographic capture–recapture sampling. Ecology 87:2925–2937
Kéry M, Royle JA (2009) Inference about species richness and community structure using species-specific occupancy models in the National Swiss Breeding Bird Survey MHB. In: Thomson DL, Cooch EG, Conroy MJ (eds) Modeling demographic processes in marked populations. Springer, New York, pp 639–656
Kéry M, Royle JA, Plattner M, Dorazio RM (2009) Species richness and occupancy estimation in communities subject to temporary emigration. Ecology 90:1279–1290
King R, Brooks SP (2001) On the Bayesian analysis of population size. Biometrika 88:317–336
King R, Brooks SP (2008) On the Bayesian estimation of a closed population size in the presence of heterogeneity and model uncertainty. Biometrics 64:816–824
King R, Brooks SP, Coulson T (2008) Analysing complex capture–recapture data in the presence of individual and temporal covariates and model uncertainty. Biometrics 64:1187–1195
Langtimm CA, Dorazio RM, Stith BM, Doyle TJ (2010) A new aerial survey design to monitor manatee abundance for Everglades restoration. J Wildl Manag (in press)
Lebreton JD, Burnham K, Clobert J, Anderson DR (1992) Modeling survival and testing biological hypotheses using marked animals: a unified approach with case studies. Ecol Monogr 62:67–118
Link WA (2003) Nonidentifiability of population size from capture–recapture data with heterogeneous detection probabilities. Biometrics 59:1123–1130
Link WA, Barker RJ (2010) Bayesian inference: with ecological applications. Academic, New York
Liu JS, Wu YN (1999) Parameter expansion for data augmentation. J Am Stat Assoc 94:1264–1274
Lunn D, Spiegelhalter D, Thomas A, Best N (2009) The BUGS project: evolution, critique and future directions (with discussion). Stat Med 28:3049–3082
MacKenzie DI, Nichols JD, Lachman GB, Droege S, Royle JA, Langtimm CA (2002) Estimating site occupancy rates when detection probabilities are less than one. Ecology 83:2248–2255
MacKenzie DI, Nichols JD, Hines JE, Knutson MG, Franklin AB (2003) Estimating site occupancy, colonization, and local extinction when a species is detected imperfectly. Ecology 84:2200–2207
Nichols JD, Karanth KU (2002) Statistical concepts: assessing spatial distributions. In: Monitoring tigers and their prey: a manual for researchers, managers, and conservationists in tropical Asia. Centre for Wildlife Studies, pp 29–38
Patil A, Huard D, Fonnesbeck CJ (2010) PyMC 2.0: Bayesian stochastic modelling in python. J Stat Softw (in press)
Pledger S (2005) The performance of mixture models in heterogeneous closed population capture–recapture. Biometrics 61:868–873
Pledger S, Pollock KH, Norris JL (2003) Open capture–recapture models with heterogeneity: I. Cormack-Jolly-Seber model. Biometrics 59:786–794
Pollock K (1982) A capture–recapture design robust to unequal probability of capture. J Wildl Manag 46:757–760
Royle JA (2006) Site occupancy models with heterogeneous detection probabilities. Biometrics 62:97–102
Royle JA (2008) Modeling individual effects in the Cormack-Jolly-Seber model: a state-space formulation. Biometrics 64:364–370
Royle JA (2009) Analysis of capture—recapture models with individual covariates using data augmentation. Biometrics 65:267–274
Royle JA, Kéry M (2007) A Bayesian state-space formulation of dynamic occupancy models. Ecology 88:1813–1823
Royle JA, Dorazio RM, Link WA (2007) Analysis of multinomial models with unknown index using data augmentation. J Comput Graph Stat 16:67–85
Royle JA, Dorazio RM (2008) Hierarchical modeling and inference in ecology: the analysis of data from populations, metapopulations and communities. Academic, San Diego
Royle JA, Gardner B (2010) Hierarchical spatial capture–recapture models for estimating density from trapping arrays. In: O'Connell AF, Nichols JD, Karanth KU (eds) Camera traps in animal ecology: methods and analyses. Springer, Berlin
Royle JA, Young KV (2008) A hierarchical model for spatial capture–recapture data. Ecology 89:2281–2289
Royle JA, Karanth KU, Gopalaswamy AM, Kumar NS (2009) Bayesian inference in camera trap studies using a class of spatial capture–recapture models. Ecology 90:3233–3244
Schwarz C, Arnason A (1996) A general methodology for the analysis of capture–recapture experiments in open populations. Biometrics 52:860–873
Schofield MR, Barker RJ (2008) A unified capture–recapture framework. J Agric Biol Environ Stat 13:458–477
Seber G (1965) A note on the multiple-recapture census. Biometrika 52:249–59
Tanner MA (1996) Tools for statistical inference: methods for the exploration of posterior distributions and likelihood functions, 3rd edn. Springer, New York
Tanner MA, Wong WH (1987) The calculation of posterior distributions by data augmentation. J Am Stat Assoc 82:528–540
Williams BK, Nichols JD, Conroy MJ (2002) Analysis and management of animal populations. Academic, San Diego
Wright JA, Barker RJ, Schofield MR, Frantz AC, Byrom AE, Gleeson DM (2009) Incorporating genotype uncertainty into mark-recapture–type models for estimating abundance using DNA samples. Biometrics 65:833–840
Acknowledgments
We thank Beth Gardner and Elise Zipkin for reviewing drafts of this manuscript. We thank Ullas Karanth (camera-trapping data) and Jim Nichols (Microtus data) for making data from their research available for our use. Use of trade, product, or firm names does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Schaub.
Appendix: Technical conditions for PX-DA
Appendix: Technical conditions for PX-DA
In applications of PX-DA to capture–recapture problems, the conventional (multinomial) model of the complete data must be expanded to account for—and to estimate—the number of non-sampling zeros in the augmented dataset of known size M. The expanded model includes an additional parameter ψ, which is related to zero inflation. Some basic conditions must be satisfied for this expanded model to be innocuous with respect to the original inference problem (Liu and Wu 1999). In this appendix, we show that the expanded model of the augmented data satisfies these conditions.
We begin with a few definitions. Let f correspond to any distribution (posterior, prior, etc.) for the observed data y, and let p denote the same for the augmented data, which we denote by (y, w). (In our applications, w is typically a vector of zeros that correspond to the all-zero capture histories.) Naturally, we desire that the posterior for the augmented data should be equivalent to the posterior based on the observed data. From Liu and Wu (1999), p(N, p | y, w) = f(N, p | y) if and only if p(N, p, ψ) “agrees with” f(N, p) in the following sense:
In other words, if the extra parameter ψ is integrated from the prior of the model of the augmented data, this yields the prior for the model of the observed data. In the RDL formulation, this integration yields a \(\hbox{U}(0,M) \times \hbox{U}(0,1)\) prior (M being some arbitrarily large integer) for the parameters (N, p), thereby satisfying Liu and Wu’s first condition. The second condition to be satisfied is that the expanded model of the augmented data, p(y, w | N, p, ψ), preserves the model of the observed data, f(y | N, p). This condition is satisfied automatically because our model of the augmented data can be considered as originating from the choice of prior on N (see “PX-DA for Model M0”), not by formulating a model that is structurally distinct from the observed-data model. Therefore, our choice of prior is sufficient to guarantee that we have satisfied the conditions of Liu and Wu (1999) and that the extra parameter ψ used in modeling the augmented data is innocuous to inference about N.
Rights and permissions
About this article
Cite this article
Royle, J.A., Dorazio, R.M. Parameter-expanded data augmentation for Bayesian analysis of capture–recapture models. J Ornithol 152 (Suppl 2), 521–537 (2012). https://doi.org/10.1007/s10336-010-0619-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-010-0619-4