Abstract
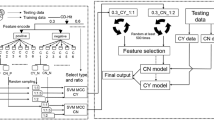
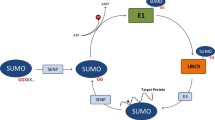
Protein sumoylation is a post-translational modification that plays an important role in a wide range of cellular processes. Small ubiquitin-related modifier (SUMO) can be covalently and reversibly conjugated to the sumoylation sites of target proteins, many of which are implicated in various human genetic disorders. The accurate prediction of protein sumoylation sites may help biomedical researchers to design their experiments and understand the molecular mechanism of protein sumoylation. In this study, a new machine learning approach has been developed for predicting sumoylation sites from protein sequence information. Random forests (RFs) and support vector machines (SVMs) were trained with the data collected from the literature. Domain-specific knowledge in terms of relevant biological features was used for input vector encoding. It was shown that RF classifier performance was affected by the sequence context of sumoylation sites, and 20 residues with the core motif ΨKXE in the middle appeared to provide enough context information for sumoylation site prediction. The RF classifiers were also found to outperform SVM models for predicting protein sumoylation sites from sequence features. The results suggest that the machine learning approach gives rise to more accurate prediction of protein sumoylation sites than the other existing methods. The accurate classifiers have been used to develop a new web server, called seeSUMO (http://bioinfo.ggc.org/seesumo/), for sequence-based prediction of protein sumoylation sites.





Similar content being viewed by others
References
Ahmad S, Sarai A (2005) PSSM-based prediction of DNA binding sites in proteins. BMC Bioinforma 6:33
Ahmad S, Gromiha MM, Sarai A (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics 20(4):477–486
Bradley A (1997) The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit 30:1145–1159
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) The proteomics protocols handbook. Humana Press, Totowa
Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8(12):947–956
Gorodkin J, Heyer LJ, Brunak S, Stormo GD (1997) Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci 13(6):583–586
Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103(1):45–50
Kawashima S, Kanehisa M (2000) AAindex: amino acid index database. Nucleic Acids Res 28(1):374
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Martin S, Wilkinson KA, Nishimune A, Henley JM (2007) Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci 8(12):948–959
Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 39(4):641–652
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567
Pu X, Guo J, Leung H, Lin Y (2007) Prediction of membrane protein types from sequences and position-specific scoring matrices. J Theor Biol 247(2):259–265
Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y (2009) Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics 9(12):3409–3412
Sarge KD, Park-Sarge OK (2009) Sumoylation and human disease pathogenesis. Trends Biochem Sci 34(4):200–205
Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18(20):6097–6100
Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D (2007) An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol 27(7):2661–2675
Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E (2004) SUMO modification of Huntingtin and Huntington’s disease pathology. Science 304(5667):100–104
Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240(4857):1285–1293
Teng S, Srivastava AK, Wang L (2010) Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics 11(Suppl 2):S5
Wang L, Brown SJ (2006a) BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res 34(Web Server issue):W243–W248
Wang L, Brown SJ (2006b) Prediction of RNA-binding residues in protein sequences using support vector machines. Conf Proc IEEE Eng Med Biol Soc 1:5830–5833
Wang L, Huang C, Yang MQ, Yang JY (2010) BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst Biol 4(Suppl 1):S3
Xu J, He Y, Qiang B, Yuan J, Peng X, Pan XM (2008) A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinforma 9:8
Xue Y, Zhou F, Fu C, Xu Y, Yao X (2006) SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res 34(Web Server issue):W254–W257
Yang SH, Galanis A, Witty J, Sharrocks AD (2006) An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J 25(21):5083–5093
Zhao J (2007) Sumoylation regulates diverse biological processes. Cell Mol Life Sci 64(23):3017–3033
Acknowledgments
This work is supported by the CSREES/USDA, under project number SC-1700355. This is technical contribution number 5913 of the Clemson Experiment Station.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Teng, S., Luo, H. & Wang, L. Predicting protein sumoylation sites from sequence features. Amino Acids 43, 447–455 (2012). https://doi.org/10.1007/s00726-011-1100-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-1100-2