Abstract
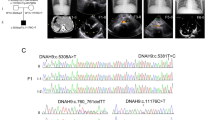
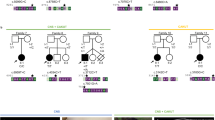
Laterality defects are heterogeneous groups of congenital malformations that arise from perturbed asymmetrical development of visceral organs. The central role of the motile cilia-generated nodal flow in breaking early embryonic symmetry is reflected in the large contribution of ciliary genes to the etiology of these disorders. In a consanguineous multiplex family with a laterality defect that resembles situs inversus totalis, and complex congenital heart disease, we combined autozygome and exome analysis to identify a novel homozygous variant in ANKS3. ANKS3 encodes a recently described ciliary protein with known interaction with other ciliary proteins, and deficiency of its zebrafish ortholog causes laterality defects. Consistent with the proposed role of the ANKS3 variant in the pathogenesis of the reported family’s phenotype, we show that the mutant RNA failed to rescue the laterality defect in anks3 morphants compared to wild-type RNA. Furthermore, we describe a new mutant anks3 line that also displays laterality defect in the homozygous state. Our study suggests a role for ANKS3 in right-left axis determination in humans.




Similar content being viewed by others
References
Alkuraya FS (2010) Autozygome decoded. Genet Med 12:765–771
Alkuraya FS (2012) Discovery of rare homozygous mutations from studies of consanguineous pedigrees. Curr Protoc Hum Genet. doi:10.1002/0471142905.hg0612s75
Alkuraya FS (2013) The application of next-generation sequencing in the autozygosity mapping of human recessive diseases. Hum Genet 132:1197–1211
Alkuraya FS (2016) Discovery of mutations for Mendelian disorders. Hum Genet 135(6):615–623. doi:10.1007/s00439-016-1664-8
Bartoloni L, Blouin J-L, Pan Y, Gehrig C, Maiti AK, Scamuffa N, Rossier C, Jorissen M, Armengot M, Meeks M (2002) Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc Natl Acad Sci 99:10282–10286
Brueckner M (2007) Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation 115:2793–2795
Cohen MS, Anderson RH, Cohen MI, Atz AM, Fogel M, Gruber PJ, Lopez L, Rome JJ, Weinberg PM (2007) Controversies, genetics, diagnostic assessment, and outcomes relating to the heterotaxy syndrome. Cardiol Young 17:29–43
Delestré L, Bakey Z, Prado C, Hoffmann S, Bihoreau M-T, Lelongt B, Gauguier D (2015) anks3 co-localises with ANKS6 in mouse renal cilia and is associated with vasopressin signaling and apoptosis in vivo in mice. PLoS One 10:e0136781
Gravesande KSVS, Omran H (2005) Primary ciliary dyskinesia: clinical presentation, diagnosis and genetics. Ann Med 37:439–449
Guichard C, Harricane M-C, Lafitte J-J, Godard P, Zaegel M, Tack V, Lalau G, Bouvagnet P (2001) Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome). Am J Hum Genet 68:1030–1035
Hoff S, Halbritter J, Epting D, Frank V, Nguyen T-MT, Van Reeuwijk J, Boehlke C, Schell C, Yasunaga T, Helmstädter M (2013) ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat Genet 45:951–956
Jacobs JP, Anderson RH, Weinberg PM, Walters HL, Tchervenkov CI, Del Duca D, Franklin RC, Aiello VD, Béland MJ, Colan SD (2007) The nomenclature, definition and classification of cardiac structures in the setting of heterotaxy. Cardiol Young 17:1–28
Kawasumi A, Nakamura T, Iwai N, Yashiro K, Saijoh Y, Belo JA, Shiratori H, Hamada H (2011) Left–right asymmetry in the level of active Nodal protein produced in the node is translated into left–right asymmetry in the lateral plate of mouse embryos. Dev Biol 353:321–330
Khoshnood B, Lelong N, Houyel L, Thieulin A-C, Jouannic J-M, Magnier S, Delezoide A-L, Magny J-F, Rambaud C, Bonnet D (2012) Prevalence, timing of diagnosis and mortality of newborns with congenital heart defects: a population-based study. Heart 98:1667–1673
Leettola CN, Knight MJ, Cascio D, Hoffman S, Bowie JU (2014) Characterization of the SAM domain of the PKD-related protein ANKS6 and its interaction with ANKS3. BMC Struct Biol 14:1
Lin AE, Ticho BS, Houde K, Westgate M-N, Holmes LB (2000) Heterotaxy: associated conditions and hospital-based prevalence in newborns. Genet Med 2:157–172
Lin AE, Krikov S, Riehle-Colarusso T, Frías JL, Belmont J, Anderka M, Geva T, Getz KD, Botto LD (2014) Laterality defects in the national birth defects prevention study (1998–2007): birth prevalence and descriptive epidemiology. Am J Med Genet Part A 164:2581–2591
Loges NT, Olbrich H, Fenske L, Mussaffi H, Horvath J, Fliegauf M, Kuhl H, Baktai G, Peterffy E, Chodhari R (2008) DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am J Hum Genet 83:547–558
Olbrich H, Häffner K, Kispert A, Völkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N (2002) Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat Genet 30:143–144
Perner B, Englert C, Bollig F (2007) The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev Biol 309:87–96
Ramachandran H, Engel C, Müller B, Dengjel J, Walz G, Yakulov TA (2015) Anks3 alters the sub-cellular localization of the Nek7 kinase. Biochem Biophys Res Commun 464:901–907
Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC (2007) p53 activation by knockdown technologies. PLoS Genet 3:e78
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi:10.1038/nmeth.2019
Shiratori H, Hamada H (2006) The left-right axis in the mouse: from origin to morphology. Development 133:2095–2104
Thisse C, Thisse B (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69
Westerfield M (1995) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 3rd edn. University of Oregon Press, Eugene, OR, p 385
Yakulov TA, Yasunaga T, Ramachandran H, Engel C, Müller B, Hoff S, Dengjel J, Lienkamp SS, Walz G (2015) Anks3 interacts with nephronophthisis proteins and is required for normal renal development. Kidney Int 87:1191–1200
Acknowledgments
We thank the study family for their enthusiastic participation. We also thank the Genotyping and Sequencing Core Facilities at KFSHRC for their technical help. This work was supported in part by a KACST Grant 13-BIO1113-20 (FSA), and by the SFB 1140 (GW).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
H. E. Shamseldin and T. A. Yakulov have contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2016_1712_MOESM1_ESM.pdf
Figure S1. (A) Clustal O alignment of zebrafish (top), human (middle) and mouse (bottom) Anks3 protein sequences. The ANKS3 protein sequence shows high degree of interspecies conservation. The ANKS3:NM_133450:exon5:c.439C>A mutation causes p.H147N change at a conserved histidine residue (red squares). The zebrafish mutant allele sa3261 contains the nonsense mutation Anks3: ENSDART00000131342:exon7:c.C888>T, which causes R272STOP amino acid change (blue square). At the beginning of each sequence the respective UniProt IDs are shown. (B) Domain predictions for the zebrafish anks3 (adapted from ENSEMBL). The R272STOP mutation causes termination of translation between the ankyrin repeats and the SAM domain. Figure S2. (A) Rescue of the anks3 heart laterality phenotypes with human wild-type and mutant anks3 mRNA. Co-injection of the anks3 splice blocking morpholino (TBM) with wild-type ANKS3 mRNA significantly reduced the observed laterality phenotypes (p < 0.01); in contrast, no recue was observed when ANKS3 mRNA carrying the mutation ANKS3:NM_133450:exon5:c.439C>A was co-injected (mutRNA). N is the number of biological repetitions; n is the total number of analyzed embryos. The error bars represent the standard deviation. (B) Injection of zebrafish embryos with either wild-type ANKS3 mRNA (anks3wt hRNA) or ANKS3 mRNA carrying the mutation ANKS3:NM_133450:exon5:c.439C>A (anks3 mut hRNA) did not increase the heart laterality defects compared to control embryos. Figure S3. (A) RT-PCR of anks3 and ef1α from wild-type and anks3 sa3261 mutant embryos. (B) Densitometric analysis of the gel bands from (A) (PDF 176 kb)
Rights and permissions
About this article
Cite this article
Shamseldin, H.E., Yakulov, T.A., Hashem, A. et al. ANKS3 is mutated in a family with autosomal recessive laterality defect. Hum Genet 135, 1233–1239 (2016). https://doi.org/10.1007/s00439-016-1712-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1712-4