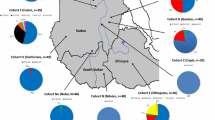
Abstract
It has been known for some 40 years that lactase production persists into adult life in some people but not in others. However, the mechanism and evolutionary significance of this variation have proved more elusive, and continue to excite the interest of investigators from different disciplines. This genetically determined trait differs in frequency worldwide and is due to cis-acting polymorphism of regulation of lactase gene expression. A single nucleotide polymorphism located 13.9 kb upstream from the lactase gene (C-13910 > T) was proposed to be the cause, and the −13910*T allele, which is widespread in Europe was found to be located on a very extended haplotype of 500 kb or more. The long region of haplotype conservation reflects a recent origin, and this, together with high frequencies, is evidence of positive selection, but also means that −13910*T might be an associated marker, rather than being causal of lactase persistence itself. Doubt about function was increased when it was shown that the original SNP did not account for lactase persistence in most African populations. However, the recent discovery that there are several other SNPs associated with lactase persistence in close proximity (within 100 bp), and that they all reside in a piece of sequence that has enhancer function in vitro, does suggest that they may each be functional, and their occurrence on different haplotype backgrounds shows that several independent mutations led to lactase persistence. Here we provide access to a database of worldwide distributions of lactase persistence and of the C-13910*T allele, as well as reviewing lactase molecular and population genetics and the role of selection in determining present day distributions of the lactase persistence phenotype.





Similar content being viewed by others
References
Anderson B, Vullo C (1994) Did malaria select for primary adult lactase deficiency? Gut 35:1487–1489
Aoki K (1986) A stochastic model of gene-culture coevolution suggested by the culture-historical hypothesis. Proc Natl Acad Sci USA 83:2929–2933
Auricchio S, Rubino A, Semenza G, Landolt M, Prader A (1963) Isolated intestinal lactase deficiency in the adult. Lancet 2:324–326
Bayoumi RA, Saha N, Salih AS, Bakkar AE, Flatz G (1981) Distribution of the lactase phenotypes in the population of the Democratic Republic of the Sudan. Hum Genet 57:279–281
Bayoumi RA, Flatz SD, Kuhnau W, Flatz G (1982) Beja and Nilotes: nomadic pastoralist groups in the Sudan with opposite distributions of the adult lactase phenotypes. Am J Phys Anthropol 58:173–178
Beja-Pereira A, Luikart G, England PR, Bradley DG, Jann OC, Bertorelle G, Chamberlain AT, Nunes TP, Metodiev S, Ferrand N, Erhardt G (2003) Gene-culture coevolution between cattle milk protein genes and human lactase genes. Nat Genet 35:311–313
Bernardes-Silva CF, Pereira AC, de Fatima Alves da Mota, Krieger JE, Laudanna AA (2007) Lactase persistence/non-persistence variants, C/T_13910 and G/A_22018, as a diagnostic tool for lactose intolerance in IBS patients. Clin Chim Acta 386:7–11
Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN (2004) Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74:1111–1120
Bloom G, Sherman PW (2005) Dairying barriers affect the distribution of lactose malabsorption. Evol Human Behav 26:301–312
Boll W, Wagner P, Mantei N (1991) Structure of the chromosomal gene and CDNAs coding for lactase-phlorizin hydrolase in humans with adult-type hypolactasia or persistence of lactase. Am J Hum Genet 48:889–902
Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD (2006a) Gata4 is essential for the maintenance of jejunal–ileal identities in the adult mouse small intestine. Mol Cell Biol 26:9060–9070
Bosse T, van Wering HM, Gielen M, Dowling LN, Fialkovich JJ, Piaseckyj CM, Gonzalez FJ, Akiyama TE, Montgomery RK, Grand RJ, Krasinski SD (2006b) Hepatocyte nuclear factor-1alpha is required for expression but dispensable for histone acetylation of the lactase-phlorizin hydrolase gene in vivo. Am J Physiol Gastrointest Liver Physiol 290:G1016–G1024
Bosse T, Fialkovich JJ, Piaseckyj CM, Beuling E, Broekman H, Grand RJ, Montgomery RK, Krasinski SD (2007) Gata4 and Hnf1alpha are partially required for the expression of specific intestinal genes during development. Am J Physiol Gastrointest Liver Physiol 292:G1302–G1314
Briet F, Pochart P, Marteau P, Flourie B, Arrigoni E, Rambaud JC (1997) Improved clinical tolerance to chronic lactose ingestion in subjects with lactose intolerance: a placebo effect? Gut 41:632–635
Buller HA, Kothe MJC, Goldman DA, Grubman SA, Sasak WV, Matsudaira PT, Montgomery RK, Grand RJ (1990) Coordinate expression of lactase-phlorizin hydrolase mRNA and enzyme levels in rat intestine during development. J Biol Chem 265:6978–6983
Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG (2007) Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA 104:3736–3741
Campbell AK, Matthews SB (2005) Darwin’s illness revealed. Postgrad Med J 81:248–251
Coelho M, Luiselli D, Bertorelle G, Lopes AI, Seixas S, stro-Bisol G, Rocha J (2005) Microsatellite variation and evolution of human lactase persistence. Hum Genet 117:329–339
Cook GC, al-Torki MT (1975) High intestinal lactase concentrations in adult Arabs in Saudi Arabia. Br Med J 3:135–136
Crittenden RG, Bennett LE (2005) Cow’s milk allergy: a complex disorder. J Am Coll Nutr 24:582S–591S
Dahlqvist A, Hammond B, Crane R, Dunphy J, Littman A (1963) Intestinal lactase deficiency and lactose intolerance in adults: preliminary report. Gastroenterology 45:488–491
Dissanyake AS, El-Munshid HA, Al-Qurain A (1990) Prevalence of primary adult lactose malabsorption in the eastern province of Saudi Arabia. Ann Saudi Med 10:598–601
Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I (2002) Identification of a variant associated with adult-type hypolactasia. Nat Genet 30:233–237
Enattah NS, Forsblom C, Rasinpera H, Tuomi T, Groop PH, Jarvela I (2004) The genetic variant of lactase persistence C (−13910) T as a risk factor for type I and II diabetes in the Finnish population. Eur J Clin Nutr 58:1319–1322
Enattah N, Pekkarinen T, Valimaki MJ, Loyttyniemi E, Jarvela I (2005a) Genetically defined adult-type hypolactasia and self-reported lactose intolerance as risk factors of osteoporosis in Finnish postmenopausal women. Eur J Clin Nutr 59:1105–1111
Enattah NS, Sulkava R, Halonen P, Kontula K, Jarvela I (2005b) Genetic variant of lactase-persistent C/T-13910 is associated with bone fractures in very old age. J Am Geriatr Soc 53:79–82
Enattah NS, Trudeau A, Pimenoff V, Maiuri L, Auricchio S, Greco L, Rossi M, Lentze M, Seo JK, Rahgozar S, Khalil I, Alifrangis M, Natah S, Groop L, Shaat N, Kozlov A, Verschubskaya G, Comas D, Bulayeva K, Mehdi SQ, Terwilliger JD, Sahi T, Savilahti E, Perola M, Sajantila A, Jarvela I, Peltonen L (2007) Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet 81:615–625
Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, El-Shanti H, Seo JK, Alifrangis M, Khalil IF, Natah A, Ali A, Natah S, Comas D, Mehdi SQ, Groop L, Vestergaard EM, Imtiaz F, Rashed MS, Meyer B, Troelsen J, Peltonen L (2008) Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet 82:57–72
Escher JC, de Koning ND, van Engen CG, Arora S, Buller HA, Montgomery RK, Grand RJ (1992) Molecular basis of lactase levels in adult humans. J Clin Invest 89:480–483
Fang R, Santiago NA, Olds LC, Sibley E (2000) The homeodomain protein Cdx2 regulates lactase gene promoter activity during enterocyte differentiation. Gastroenterology 118:115–127
Fang R, Olds LC, Santiago NA, Sibley E (2001) GATA family transcription factors activate lactase gene promoter in intestinal Caco-2 cells. Am J Physiol Gastrointest Liver Physiol 280:G58–G67
Ferguson A, Maxwell J (1967) Genetic aetiology of lactose intolerance. Lancet 2:188–190
Flatz G (1984) Gene dosage effect on intestinal lactase activity demonstrated in vivo. Am J Hum Genet 36:306–310
Flatz G (1987) Genetics of lactose digestion in humans. In: Harris H, Hirschhorn K (eds) Advances in human genetics, vol 16. Plenum Press, New York, pp 1–77
Flatz G, Rotthauwe HW (1973) Lactose nutrition and natural selection. Lancet 2:76–77
Gueguen L, Pointillart A (2000) The bioavailability of dietary calcium. J Am Coll Nutr 19:119S–136S
HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796
Harju M (2003) Chromatographic and enzymatic removal of lactose from milk, IDF World Dairy Summit, Bruges, BELGIQUE (09/2003) 2004, no 389, pp 4–8
Harvey CB, Pratt WS, Islam I, Whitehouse DB, Swallow DM (1995) DNA polymorphisms in the lactase gene. Linkage disequilibrium across the 70-kb region. Eur J Hum Genet 3:27–41
Harvey CB, Hollox EJ, Poulter M, Wang Y, Rossi M, Auricchio S, Iqbal TH, Cooper BT, Barton R, Sarner M, Korpela R, Swallow DM (1998) Lactase haplotype frequencies in Caucasians: association with the lactase persistence/non-persistence polymorphism. Ann Hum Genet 62(Pt 3):215–223
Hauri HP, Sterchi EE, Bienz D, Fransen JA, Marxer A (1985) Expression and intracellular transport of microvillus membrane hydrolases in human intestinal epithelial cells. J Cell Biol 101:838–851
Hertzler SR, Savaiano DA (1996) Colonic adaptation to daily lactose feeding in lactose maldigesters reduces lactose intolerance. Am J Clin Nutr 64:232–236
Hertzler SR, Savaiano DA, Levitt MD (1997) Fecal hydrogen production and consumption measurements—response to daily lactose ingestion by lactose maldigesters. Dig Dis Sci 42:348–353
Ho MW, Povey S, Swallow DM (1982) Lactase polymorphism in adult British natives: estimating allele frequencies by enzyme assays in autopsy samples. Am J Hum Genet 34:650–657
Hogenauer C, Hammer HF, Mellitzer K, Renner W, Krejs GJ, Toplak H (2005) Evaluation of a new DNA test compared with the lactose hydrogen breath test for the diagnosis of lactase non-persistence. Eur J Gastroenterol Hepatol 17:371–376
Holden C, Mace R (1997) Phylogenetic analysis of the evolution of lactose digestion in adults. Hum Biol 69:605–628
Hollox EJ, Poulter M, Wang Y, Krause A, Swallow DM (1999) Common polymorphism in a highly variable region upstream of the human lactase gene affects DNA–protein interactions. Eur J Hum Genet 7:791–800
Hollox EJ, Poulter M, Zvarik M, Ferak V, Krause A, Jenkins T, Saha N, Kozlov AI, Swallow DM (2001) Lactase haplotype diversity in the Old World. Am J Hum Genet 68:160–172
Imtiaz F, Savilahti E, Sarnesto A, Trabzuni D, Al-Kahtani K, Kagevi I, Rashed MS, Meyer BF, Jarvela I (2007) The T/G 13915 variant upstream of the lactase gene (LCT) is the founder allele of lactase persistence in an urban Saudi population. J Med Genet 44:e89
Ingram CJE (2008) The evolutionary genetics of lactase persistence in Africa and the Middle East. Ph.D. thesis University of London
Ingram CJE, Elamin MF, Mulcare CA, Weale ME, Tarekegn A, Raga TO, Bekele E, Elamin FM, Thomas MG, Bradman N, Swallow DM (2007) A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum Genet 120:779–788
Jacob R, Brewer C, Fransen JA, Naim HY (1994) Transport, function, and sorting of lactase-phlorizin hydrolase in Madin–Darby canine kidney cells. J Biol Chem 269:2712–2721
Jacob R, Bulleid NJ, Naim HY (1995) Folding of human intestinal lactase-phlorizin hydrolase. J Biol Chem 270:18678–18684
Jacob R, Radebach I, Wuthrich M, Grunberg J, Sterchi EE, Naim HY (1996) Maturation of human intestinal lactase-phlorizin hydrolase: generation of the brush border form of the enzyme involves at least two proteolytic cleavage steps. Eur J Biochem 236:789–795
Jacob R, Peters K, Naim HY (2002) The prosequence of human lactase-phlorizin hydrolase modulates the folding of the mature enzyme. J Biol Chem 277:8217–8225
Kerber M, Oberkanins C, Kriegshauser G, Kollerits B, senbach-Glaninger A, Fuchs D, Ledochowski M (2007) Hydrogen breath testing versus LCT genotyping for the diagnosis of lactose intolerance: a matter of age? Clin Chim Acta 383:91–96
Krasinski SD, van Wering HM, Tannemaat MR, Grand RJ (2001) Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am J Physiol Gastrointest Liver Physiol 281:G69–G84
Lacey SW, Naim HY, Magness RR, Gething M-J, Sambrook JF (1994) Expression of lactase-phlorizin hydrolase in sheep is regulated at the RNA level. Biochem J 302:929–935
Larsson SC, Orsini N, Wolk A (2006) Milk, milk products and lactose intake and ovarian cancer risk: a meta-analysis of epidemiological studies. Int J Cancer 118:431–441
Lee SY, Wang Z, Lin CK, Contag CH, Olds LC, Cooper AD, Sibley E (2002) Regulation of intestine-specific spatiotemporal expression by the rat lactase promoter. J Biol Chem 277:13099–13105
Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, Troelsen JT (2005) T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet 14:3945–3953
Lloyd M, Mevissen G, Fischer M, Olsen W, Goodspeed D, Genini M, Boll W, Semenza G, Mantei N (1992) Regulation of intestinal lactase in adult hypolactasia. J Clin Invest 89:524–529
Maiuri L, Raia V, Potter J, Swallow DM, Ho MW, Fiocca R, Finzi G, Cornaggia M, Capella C, Quaroni A, Auricchio S (1991) Mosaic pattern of lactase expression in villous enterocytes in human adult-type hypolactasia. Gastroenterology 100:359–369
Maiuri L, Rossi M, Raia V, Garipoli V, Hughes LA, Swallow D, Noren O, Sjostrom H, Auricchio S (1994) Mosaic regulation of lactase in human adult-type hypolactasia. Gastroenterology 107:54–60
Matthews SB, Waud JP, Roberts AG, Campbell AK (2005) Systemic lactose intolerance: a new perspective on an old problem. Postgrad Med J 81:167–173
McCracken RD (1971) Lactase deficiency—example of dietary evolution. Curr Anthropol 12:479
Meloni T, Colombo C, Ruggiu G, Dessena M, Meloni GF (1998) Primary lactase deficiency and past malarial endemicity in Sardinia. Ital J Gastroenterol Hepatol 30:490–493
Meloni GF, Colombo C, La VC, Ruggiu G, Mannazzu MC, Ambrosini G, Cherchi PL (1999) Lactose absorption in patients with ovarian cancer. Am J Epidemiol 150:183–186
Meloni GF, Colombo C, La VC, Pacifico A, Tomasi P, Ogana A, Marinaro AM, Meloni T (2001) High prevalence of lactose absorbers in Northern Sardinian patients with type 1 and type 2 diabetes mellitus. Am J Clin Nutr 73:582–585
Metneki J, Cziezel A, Flatz SD, Flatz G (1984) A study of lactose absorption capacity in twins. Hum Genet 67:296–300
Mitchelmore C, Troelsen JT, Spodsberg N, Sjostrom H, Noren O (2000) Interaction between the homeodomain proteins Cdx2 and HNF1alpha mediates expression of the lactase-phlorizin hydrolase gene. Biochem J 346(Pt 2):529–535
Mulcare (2006) The evolution of the lactase persistence phenotype. Ph.D. thesis University of London, London, UK
Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, Swallow DM, Bradman N, Thomas MG (2004) The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet 74:1102–1110
Naim HY, Lentze MJ (1992) Impact of O-glycosylation on the function of human intestinal lactase-phlorizin hydrolase. Characterization of glycoforms varying in enzyme activity and localization of O-glycoside addition. J Biol Chem 267:25494–25504
Newcomer AD, McGill DB, Thomas PJ, Hofmann AF (1975) Prospective comparison of indirect methods for detecting lactase deficiency. N Engl J Med 293:1232–1236
Obermayer-Pietsch BM, Bonelli CM, Walter DE, Kuhn RJ, Fahrleitner-Pammer A, Berghold A, Goessler W, Stepan V, Dobnig H, Leb G, Renner W (2004) Genetic predisposition for adult lactose intolerance and relation to diet, bone density, and bone fractures. J Bone Miner Res 19:42–47
Olds LC, Sibley E (2003) Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet 12:2333–2340
Peuhkuri (2000) Lactose, lactase, and bowel disorders, Ph.D. thesis, University of Helsinki, Finland
Peuhkuri K, Vapaatalo H, Korpela R, Teuri U (2000) Lactose intolerance—a confusing clinical diagnosis. Am J Clin Nutr 71:600–602
Poulter M, Hollox E, Harvey CB, Mulcare C, Peuhkuri K, Kajander K, Sarner M, Korpela R, Swallow DM (2003) The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet 67:298–311
Rasinpera H, Savilahti E, Enattah NS, Kuokkanen M, Totterman N, Lindahl H, Jarvela I, Kolho KL (2004) A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut 53:1571–1576
Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES (2006) Positive natural selection in the human lineage. Science 312:1614–1620
Sahi T (1974) The inheritance of selective adult-type lactose malabsorption. Scand J Gastroenterol 9:1–73
Saltzman JR, Russell RM, Golner B, Barakat S, Dallal GE, Goldin BR (1999) A randomized trial of Lactobacillus acidophilus BG2FO4 to treat lactose intolerance. Am J Clin Nutr 69:140–146
Scrimshaw NS, Murray EB (1988) The acceptability of milk and milk products in populations with a high prevalence of lactose intolerance. Am J Clin Nutr 48:1079–1159
Sebastio G, Villa M, Sartorio R, Guzzetta V, Poggi V, Auricchio S, Boll W, Mantei N, Semenza G (1989) Control of lactase in human adult-type hypolactasia and in weaning rabbits and rats. Am J Hum Genet 45:489–497
Simoons FJ (1970) Primary lactose intolerance and the milking habit: a problem in biological and cultural interrelations, II. A culture historical hypothesis. Am J Dig Dis 15:695–710
Simoons FJ (1978) The geographic hypothesis and lactose malabsorption—a weighing of the evidence. Dig Dis 23:963–980
Snook CR, Mahmoud JN, Chang WP (1976) Lactose tolerance in adult Jordanian Arabs. Trop Geogr Med 28:333–335
Spodsberg N, Troelsen JT, Carlsson P, Enerback S, Sjostrom H, Noren O (1999) Transcriptional regulation of pig lactase-phlorizin hydrolase. Involvement of HNF-1 and FREACs. Gastroenterology 116:842–854
Sterchi E, Mills P, Fransen J, Hauri H, Lentze M, Naim H, Ginsel L, Bond J (1990) Biogenesis of intestinal lactase-phlorizin hydrolase in adults with lactose intolerance. Evidence for reduced biosynthesis and slowed-down maturation in enterocytes. J Clin Invest 86:1329–1337
Suarez FL, Savaiano D, Arbisi P, Levitt MD (1997) Tolerance to the daily ingestion of two cups of milk by individuals claiming lactose intolerance. Am J Clin Nutr 65:1502–1506
Swallow DM (2006) DNA test for hypolactasia premature. Gut 55:131–132
Swallow DM, Hollox EJ (2000) The genetic polymorphism of intestinal lactase activity in adult humans. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited disease. McGraw-Hill, New York, pp 1651–1662
Tag CG, Schifflers MC, Mohnen M, Gressner AM, Weiskirchen R (2007) A novel proximal −13914G > A base replacement in the vicinity of the common-13910T/C lactase gene variation results in an atypical LightCycler melting curve in testing with the MutaREAL lactase test. Clin Chem 53:146–148
Tag CG, Oberkanins C, Kriegshauser G, Ingram CJ, Swallow DM, Gressner AM, Ledochowski M, Weiskirchen R (2008) Evaluation of a novel reverse-hybridization StripAssay for typing DNA variants useful in diagnosis of adult-type hypolactasia. Clin Chim Acta 392:58–62
Thacher TD, Fischer PR, Pettifor JM, Lawson JO, Isichei CO, Reading JC, Chan GM (1999) A comparison of calcium, vitamin D, or both for nutritional rickets in Nigerian children. N Engl J Med 341:563–568
Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, Ibrahim M, Omar SA, Lema G, Nyambo TB, Ghori J, Bumpstead S, Pritchard JK, Wray GA, Deloukas P (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39:31–40
Troelsen JT, Mehlum A, Olsen J, Spodsberg N, Hansen GH, Prydz H, Noren O, Sjostrom H (1994) 1 kb of the lactase-phlorhizin hydrolase promoter directs post-weaning decline and small intestinal-specific expression in transgenic mice. FEBS Lett 342:291–296
Troelsen JT, Mitchelmore C, Spodsberg N, Jensen AM, Noren O, Sjostrom H (1997) Regulation of lactase-phlorizin hydrolase gene expression by the caudal-related homoeodomain protein Cdx-2. Biochem J 322(Pt 3):833–838
Troelsen JT, Olsen J, Moller J, Sjostrom H (2003) An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology 125:1686–1694
van Wering HM, Bosse T, Musters A, de Jong E, de Jong N, Hogen Esch CE, Boudreau F, Swain GP, Dowling LN, Montgomery RK, Grand RJ, Krasinski SD (2004) Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physiol Gastrointest Liver Physiol 287(4):G899–909
Vesa TH, Marteau P, Korpela R (2000) Lactose intolerance. J Am Coll Nutr 19:165S–175S
Villako K, Maaroos H (1994) Clinical picture of hypolactasia and lactose intolerance. Scand J Gastroenterol Suppl 202:36–54
Wang Y, Harvey CB, Pratt WS, Sams VR, Sarner M, Rossi M, Auricchio S, Swallow DM (1995) The lactase persistence/non-persistence polymorphism is controlled by a cis-acting element. Hum Mol Genet 4:657–662
Wang Z, Maravelias C, Sibley E (2006) Lactase gene promoter fragments mediate differential spatial and temporal expression patterns in transgenic mice. DNA Cell Biol 25:215–222
Weiskirchen, Tag CG, Mengsteab S, Gressner AM, Ingram CJE, Swallow DM (2007) Pitfalls in LightCycler Diagnosis of the single-nucleotide polymorphism 13.9 kb upstream of the lactase gene that is associated with adult-type hypolactasia
Witte J, Lloyd M, Lorenzsonn V, Korsmo H, Olsen W (1990) The biosynthetic basis of adult lactase deficiency. J Clin Invest 86:1338–1342
Acknowledgments
CJEI and CAM were funded by BBSRC CASE studentships and YI was funded by UCL Graduate school, UCL ORS and B’nai B’rith/Leo Baeck London Lodge scholarships. We thank Neil Bradman, The Centre for Genetic Anthropology, UCL, for access to samples and Melford Charitable Trust for funding.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary figure 1a
Interpolated maps of the ‘old world’ showing the distribution of (a) lactase persistence data taken from the literature (Supplementary data Table 1), (b) -13910*T distribution (c) lactase persistence frequency predicted from -13910*T distribution, using the data collection to be found in Supplementary data Table 3. Maps were generated using PYNGL (http://www.pyngl.ucar.edu). Only includes individuals over 12 years of age, who are unrelated, and literature for which the original publications have been located and checked. Articles in which there was clear selection bias, and recent immigrant populations are excluded, but the data can be found in Supplementary data Table 1. The Americas are excluded from all maps because of the paucity of data. Most data are obtained from lactose tolerance tests using either breath hydrogen or blood glucose, though in some cases enzyme assay data were available. Locations were either as described precisely in the publication, or taken from capital cities or central points of a country or region where precise location is not mentioned. Where more than one data set was available weighted averages of the data were taken. Predicted frequency taken to be p2+ 2pq, where p is the frequency of -13910*T. Data points are shown as dots. It should be noted that the interpolation is inaccurate where there are few data points (PDF 809 kb)
Supplementary figure 1b
See supplementary figure 1a for legend (PDF 752 kb)
Supplementary figure 1c
See supplementary figure 1a for legend (PDF 766 kb)
439_2008_593_MOESM4_ESM.pdf
Primary source literature references of lactase persistence and lactose tolerance data used for maps depicting geographic distribution. Columns show numbers of people tested, country of origin, ethnic group, test method, and whether or not the data fulfilled all the criteria for inclusion (original reference found and checked; unrelated individuals; age 12 or more; unbiased selection criteria - e.g. not selected from patients with diarrhoea). Reasons for non-inclusion are shown in the notes. In those cases where children or family members were individually identifiable, they were excluded from the data sets and this is reflected in the numbers given. Recent immigrant populations are excluded from the maps shown in the review article. Locations (longitude and latitude) were either as described precisely in the original publication, or taken from capital cities or central points of a country in those cases that the precise location is not mentioned. Data included only in review articles were not used. Reviews searched for source references include: Flatz, (1987); Scrimshaw and Murray, (1988); Swallow and Hollox, (1999) (PDF 39.5 kb)
439_2008_593_MOESM5_ESM.pdf
Estimates of lactase persistence frequency in different countries obtained by adjusting for population census size (taken from CIA data; World Fact Book http://www.cia.gov) (PDF 29.0 kb)
439_2008_593_MOESM6_ESM.pdf
Literature and own frequency data for -13910*T. Data taken from SNP typing tests as well as from resequencing. Predicted lactase persistence frequency attributable to this allele taken to be p2 + 2pq (PDF 21.7 kb)
Rights and permissions
About this article
Cite this article
Ingram, C.J.E., Mulcare, C.A., Itan, Y. et al. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet 124, 579–591 (2009). https://doi.org/10.1007/s00439-008-0593-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-008-0593-6