Abstract
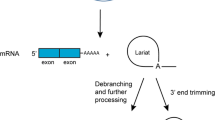
Intron lariat RNAs, created by pre-mRNA splicing, are sources of information on gene expression and structure. Although produced equivalently to their corresponding mRNAs, the vast majority of intron lariat RNAs are rapidly degraded. However, their levels are enhanced in cells deficient for RNA debranching enzyme, which catalyzes linearization of these RNAs, the rate-limiting step in their degradation. Furthermore, RNA lariats are resistant to degradation by the 3′ exonuclease polynucleotide phosphorylase (PNPase), providing a means to enrich for lariat RNAs. Working with the yeast Saccharomyces cerevisiae as a model organism, our goal was to develop novel combinations of methods to enhance the use of intron lariat RNAs as objects of study. Using RT-PCR assays developed for detecting and quantifying specific lariat RNAs, we demonstrate the resistance of RNA lariats to degradation by PNPase and their sensitivity to cleavage by RNA debranching enzyme. We also employ sequential treatments with these two enzymes to produce characteristic effects on linear and lariat RNAs. We establish the utility of the methods for analyzing RNA debranching enzyme variants and in vitro debranching reactions and discuss several possible applications, including measuring relative rates of transcription and combining these methods with non-gene-specific RNA sequencing as a novel approach for genome annotation. In summary, enzymatic treatments that produce characteristic effects on linear and lariat RNAs, combined with RT-PCR or RNA sequencing, can be powerful tools to advance studies on gene expression, alternative splicing, and any process that depends on the RNA debranching enzyme.







Similar content being viewed by others
References
Abelson J (2008) Is the spliceosome a ribonucleoprotein enzyme? Nat Struct Mol Biol 15(12):1235–1237
Alani E, Cao L, Kleckner N (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116(4):541–545
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2003) Current protocols in molecular biology on CD-ROM. Current Protocols Inc., Brooklyn
Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC (2007) Mammalian mirtron genes. Mol Cell 28(2):328–336
Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK (2009) Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog 5(5):e1000437
Chapman KB, Boeke JD (1991) Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65:483–492
Cheng Z, Menees TM (2004) RNA branching and debranching in the yeast retrovirus-like element Ty1. Science 303:240–243
Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, Robertson AJ, Perkins AC, Bruce SJ, Lee CC, Ranade SS, Peckham HE, Manning JM, McKernan KJ, Grimmond SM (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5(7):613–619
Conklin JF, Goldman A, Lopez AJ (2005) Stabilization and analysis of intron lariats in vivo. Methods 37(4):368–375
Coombes CE, Boeke JD (2005) An evaluation of detection methods for large lariat RNAs. RNA 11(3):323–331
Daniels DL, Michels WJ Jr, Pyle AM (1996) Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J Mol Biol 256(1):31–49
Engleman A (2010) Reverse transcription and integration. In: Kurth R, Bannert N (eds) Retroviruses: molecular biology, genomics, and pathogenesis. Caister Academic Press, Norfolk, pp 129–159
Falaleeva MV, Chetverina HV, Ugarov VI, Uzlova EA, Chetverin AB (2008) Factors influencing RNA degradation by Thermus thermophilus polynucleotide phosphorylase. FEBS J 275(9):2214–2226
Filipowicz W, Pogacic V (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 14(3):319–327
Gallwitz D, Sures I (1980) Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 77(5):2546–2550
Gao K, Masuda A, Matsuura T, Ohno K (2008) Human branch point consensus sequence is yUnAy. Nucleic Acids Res 36(7):2257–2267
Garcia-Martinez J, Aranda A, Perez-Ortin JE (2004) Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 15(2):303–313
Goff SP (2007) Retroviridae: the retroviruses and their replication. In: Knipe DM, Howley PM (eds) Fields virology. Lipincott, Williams, and Wilkins, Philadelphia, pp 1999–2069
Gray M, Kupiec M, Honigberg SM (2004) Site-specific genomic (SSG) and random domain-localized (RDL) mutagenesis in yeast. BMC Biotechnol 4:7
Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Devine SE (2003) Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164(3):867–879
Guarneros G, Portier C (1990) Different specificities of ribonuclease II and polynucleotide phosphorylase in 3′mRNA decay. Biochimie 72(11):771–777
Hallegger M, Llorian M, Smith CW (2010) Alternative splicing: global insights. FEBS J 277(4):856–866
Hill JE, Myers AM, Koerner TJ, Tzagoloff A (1986) Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2(3):163–167
Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95(5):717–728
Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S (2005) Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res 15(5):641–654
Juneau K, Palm C, Miranda M, Davis RW (2007) High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci USA 104(5):1522–1527
Kaiser C, Michaelis S, Mitchell A (1994) Methods in yeast genetics. CSHL Press, Cold Spring Harbor
Karst SM, Rutz M-L, Menees TM (2000) The yeast retrotransposons Ty1 and Ty3 required the RNA lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. BioChem Biophys Res Comm 268:112–117
Kataoka N, Fujita M, Ohno M (2009) Functional association of the Microprocessor complex with the spliceosome. Mol Cell Biol 29(12):3243–3254
Khalid MF, Damha MJ, Shuman S, Schwer B (2005) Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res 33(19):6349–6360
Kim YK, Kim VN (2007) Processing of intronic microRNAs. EMBO J 26(3):775–783
Kiss T, Fayet E, Jady BE, Richard P, Weber M (2006) Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol 71:407–417
Lestrade L, Weber MJ (2006) snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34(database issue):D158–162
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133(3):523–536
Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H (1999) Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153(4):1535–1546
Lopez PJ, Seraphin B (2000) YIDB: the Yeast Intron DataBase. Nucleic Acids Res 28(1):85–86
Martin A, Schneider S, Schwer B (2002) Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277(20):17743–17750
McLaren RS, Newbury SF, Dance GS, Causton HC, Higgins CF (1991) mRNA degradation by processive 3’–5’ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol 221(1):81–95
Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, Martin DE, Tresch A, Cramer P (2011) Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7:458
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320(5881):1344–1349
Nam K, Hudson RH, Chapman KB, Ganeshan K, Damha MJ, Boeke JD (1994) Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem 269(32):20613–20621
Ng R, Abelson J (1980) Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 77(7):3912–3916
Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463(7280):457–463
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130(1):89–100
Ooi SL, Samarsky DA, Fournier MJ, Boeke JD (1998) Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4(9):1096–1110
Ooi SL, Dann C 3rd, Nam K, Leahy DJ, Damha MJ, Boeke JD (2001) RNA lariat debranching enzyme. Methods Enzymol 342:233–248
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40(12):1413–1415
Pastuszak AW, Joachimiak MP, Blanchette M, Rio DC, Brenner SE, Frankel AD (2010) An SF1 affinity model to identify branch point sequences in human introns. Nucleic Acids Res 39(6):2344–2356
Pelechano V, Perez-Ortin JE (2010) There is a steady-state transcriptome in exponentially growing yeast cells. Yeast 27(7):413–422
Pratico ED, Silverman SK (2007) Ty1 reverse transcriptase does not read through the proposed 2’, 5’-branched retrotransposition intermediate in vitro. RNA 13(9):1528–1536
Preker PJ, Guthrie C (2006) Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA 12(6):994–1006
Preker PJ, Kim KS, Guthrie C (2002) Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8(8):969–980
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14(10A):1902–1910
Rodriguez-Navarro S, Strasser K, Hurt E (2002) An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast. EMBO Rep 3(5):438–442
Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448(7149):83–86
Salem LA, Boucher CL, Menees TM (2003) Relationship between RNA lariat debranching and yeast Ty1 element retrotransposition. J Virol 77:12795–12806
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122(1):19–27
Smale ST (2009). Nuclear run-on assay. Cold Spring Harb Protoc 2009(11): pdb prot5329
Smith DJ, Query CC, Konarska MM (2008) “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell 30(6):657–666
Spingola M, Grate L, Haussler D, Ares M Jr (1999) Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5(2):221–234
Storici F, Lewis LK, Resnick MA (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol 19(8):773–776
Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, Schmidt D, O’Keeffe S, Haas S, Vingron M, Lehrach H, Yaspo ML (2008) A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321(5891):956–960
Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34(8):e63
Tang GQ, Maxwell ES (2008) Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res 18(1):104–112
Vincent HA, Deutscher MP (2006) Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem 281(40):29769–29775
Vogel J, Borner T (2002) Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J 21(14):3794–3803
Vogel J, Hess WR, Borner T (1997) Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res 25(10):2030–2031
Wahl MC, Will CL, Luhrmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136(4):701–718
Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453(7199):1239–1243
Ye Y, De Leon J, Yokoyama N, Naidu Y, Camerini D (2005) DBR1 siRNA inhibition of HIV-1 replication. Retrovirology 2:63
Zhang Z, Hesselberth JR, Fields S (2007) Genome-wide identification of spliced introns using a tiling microarray. Genome Res 17(4):503–509
Acknowledgments
We thank Beate Schwer for the gift of pET16b-DBR1 and Haoping Liu for yeast sigma strain 10560-23C. Support was provided by National Science Foundation, the University of Missouri Research Board, and the University of Missouri-Kansas City School of Biological Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Aguilera.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheng, Z., Menees, T.M. RNA splicing and debranching viewed through analysis of RNA lariats. Mol Genet Genomics 286, 395–410 (2011). https://doi.org/10.1007/s00438-011-0635-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-011-0635-y