Abstract
We report the integration of a type II restriction-methylase, mFokI, into the tobacco chloroplast genome and we demonstrate that the introduced enzyme effectively directs the methylation of its target sequence in vivo and does not affect maternal inheritance. We further report the transformation of tobacco with an E. coli dcm methylase targeted to plastids and we demonstrate efficient cytosine methylation of the plastid genome. Both adenosine methylation of FokI sites and cytosine methylation of dcm sites appeared phenotypically neutral. The ability to tolerate such plastid genome methylation is a pre-requisite for a proposed plant transgene containment system. In such a system, a chloroplast located, maternally inherited restriction methylase would provide protection from a nuclear-encoded, plastid targeted restriction endonuclease. As plastids are not paternally inherited in most crop species, pollen from such plants would carry the endonuclease transgene but not the corresponding methylase; the consequence of this should be containment of all nuclear transgenes, as pollination will only be viable in crosses to the appropriate transplastomic maternal background.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The presence of low levels of methyl cytosine and adenosine within plastid DNA of liverwort (Takio et al. 1994), pea (Ohta et al. 1991), sycamore (Ngernprasirtsiri et al. 1988), maize (Gauly and Kössel 1989) and tomato (Kobayashi et al. 1990) have been reported, and an association with plant development has been proposed (Kobayashi et al. 1990). However, other groups have not always been able to duplicate such results (Marano and Carrillo 1991) and it has not been possible to detect reported methylation using bisulfite sequencing (Fojtová et al. 2001). This has led to the proposal that much of the reported methylation may be an artifact due to the use of inappropriate restriction enzymes for its detection, primarily EcoRII (Fojtová et al. 2001).
An exception to this is the chloroplast DNA of the unicellular green alga, Chlamydomonas reinhardtii, where extensive cytosine methylation is found (Sager 1972). There are two Chlamydomonas mating types, and hypermethylation of chloroplast DNA is associated with the maternal mating type (mating type plus, mt+; Sager and Ramanis 1973; Burton et al. 1979). Under normal conditions, progeny inherit chloroplast DNA from the mt+ parent and that from the paternal mating type (mating type minus; mt−) is lost. Methylation of the chloroplast DNA is directed by a nuclear encoded, chloroplast targeted methylase, and constitutive expression of this gene in transgenic mt− cells leads to methylation of the paternal chloroplast genome and a high frequency of chloroplasts being inherited from the paternal parent (Nishiyama et al. 2004). It was originally proposed that this uniparental inheritance could be brought about by a restriction/modification system acting to protect maternal and to digest non-methylated paternal chloroplast DNA (Sager and Lane 1972). However, more recent studies demonstrate that methylation is not necessary for protection from nucleases and support a model in which it promotes differential chloroplast DNA replication in germinating zygotes (Umen and Goodenough 2001).
In most crop plants, plastids are only inherited maternally; this is primarily a result of exclusion from the pollen generative cell, although examples of rare paternal transmission have been reported (Ruf et al. 2007; Svab and Maliga 2007). We postulated that bacterial restriction/methylation systems could be adapted and used to engineer transgenic plants in order to provide a mechanism for preventing transgenes from out crossing to non-transgenic crops or wild relatives. Restriction-modification systems are believed to have evolved as a natural defence mechanism to protect bacteria against invading bacteriophages. The restriction endonuclease recognises short, specific nucleotide sequences in foreign DNA entering the bacterial cell and cuts at or near the recognised site. At the same time, the corresponding restriction methylase provides protection for the host’s DNA by adding a methyl group to the same target sequence. Three classes of restriction/modification systems are recognised in bacteria but only the type II system has the sequence specific methylation and sequence specific cleavage activities as separate enzyme functions (reviewed in Murray 2000; Bickle and Kruger 1993). Such systems have the potential to be modified to provide a method for transgene containment in plants. If a gene encoding a bacterial restriction endonuclease were to be equipped with a chloroplast import sequence and used for plant transformation, then the endonuclease would be anticipated to cleave the plastid genome at its target sites–destroying the chloroplast and leading to cell death. However, integration of a sequence specific methylase transgene into the plastid genome should result in methylation at target sites, conferring resistance to the imported nuclease, provided that all sites are methylated and that methylation of plastid DNA is tolerated by the plant. The potential number of such containment systems is theoretically in the hundreds, only being limited by the number of restriction endonuclease/methylase pairs available with recognition sites within the chloroplast genome.
The FokI restriction/modification system is the best characterised member of a small subset of the type II group in which the endonuclease activity is conferred by a monomer rather than two polypeptides acting as a homodimer (Kita et al. 1989). We anticipated that a monomer might be more readily taken up by the chloroplast import apparatus and for this reason the FokI system was chosen for engineering for plant expression. The FokI recognition sequence is GGATG which occurs 237 times in the tobacco chloroplast genome. Protection from digestion by FokI endonuclease (RFokI) is conferred by the FokI methylase (MFokI) which methylates the central adenosine within the recognition sequence (Sugisaki et al. 1989). As a first step in the creation of an outcross isolation system, we report the generation of homoplastomic plants carrying the MFokI transgene and demonstrate methylation at most, but not all, of the cognate sites. No deleterious effects on plant growth or plastid inheritance were observed.
To test whether cytosine methylation of the plastid genome could also be tolerated by the plants we transformed wild type plants with an E. coli dcm methylase transgene (Marinus and Morris 1973) which we equipped with a plastid import leader sequence. Extensive methylation of target sites was observed and again, no obvious deleterious effects on plant growth were seen. The demonstration of the successful insertion of a restriction methylase into the chloroplast genome and the ability of transgenic plants to tolerate extensive methylation of both adenosines and cytosines within their plastid genome is a critical first step in engineering restriction enzyme based containment systems.
Materials and methods
Plant material
Tobacco (N. tabacum cv. Petit Havana) plants were grown in MSR3 media (Waters et al. 2004). Transplastomic lines were rooted and propagated on MSR3 supplemented with 500 μg ml−1 of spectinomycin. Nuclear transformants were grown on the same media supplemented with 50 μg ml−1 kanamycin.
Construction of the chloroplast transformation vector
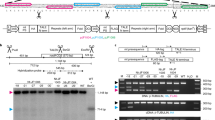
The plastid transformation vector pFaadMe2t was based upon pFaadAI ΔSN, kindly provided by Eibl et al. (1999). The pFaadAI ΔSN vector includes a sequence corresponding to nucleotides 114,279–116,171 of the tobacco plastid genome for facilitating integration through homologous recombination, an aadA resistance marker under the control of the 16S-rDNA promoter, a 25 bp ribosome binding site synthesised to match the corresponding sequence of the tobacco rbcL gene, and the 3′- end (450 bp) of the C. reinhardtii rbcL gene. In order to create pFaadMet2 the mFokI gene was amplified by PCR from the bacterium F. okeanokoites using the oligonucleotides 5′-MFokI CCACCGCGGAGGAGGTGAAAATATGAGATTTATTGG and 3′-MFokI GGAGCATGCGATTTATTGGGAGCAAAG and the amplified product ligated into the pTAg PCR cloning vector (R&D Systems, UK). The resulting plasmid was subsequently digested with SphI to release a fragment encoding the the methylase as a 1.9 kb fragment which was then ligated into the SphI site of pFaadΔSN between the aadA gene and the 3′end of rbcL to create pFaadMet2 (Fig. 1a). E. coli clones were functionally selected through the ability of plasmid DNA to withstand digestion with FokI endonuclease.
Chloroplast transformation with the FokI methylase. The transformation vector pFaadMet2 targets transgene insertion to the small single copy region at a site proximal to the ndhF gene (a). Insertion at this site results in an increase in size of 3,278 bp for the BglII fragment containing the ndhF probe sequence (b). Following Southern analysis, the absence of the smaller band derived from the wild type (WT) plastome indicates that all six transplastomic plants are homoplastomic (lanes 1–6)
Generation of transplastomic plants
Leaves were placed with abaxial side up on RMOP medium (Svab et al. 1990) in a petri dish. Preparation of and bombardment with particles was performed as described by Bock (1998) except gold particles (0.6 μm) were prepared instead of tungsten. Biolistics was carried out using the DuPont PDS1000He biolistic gun and 1,100 psi rupture disks (Bio-Rad). After bombardment the leaves were incubated at 25°C and 1 to 2 days later the leaves were cut into sections (5 mm × 5 mm) and transferred to RMOP medium containing 500 μg ml−1 of spectinomycin dyhydrochlororide. Green calli that formed on leaf segments were subcultured onto the same selective medium. Shooting leaves were then re-callused a second time and subsequent shoots rooted on MS medium.
Construction of dcm methylase binary vectors and plant transformation
Plasmid pBSRBCS contains the petunia SSU611 ribulose bisphosphate carboxylase small subunit (rbcS) chloroplast targeting sequence (Dean et al. 1987) cloned into pBluescript II SK+ (Stratagene, UK). This sequence is derived from plasmid pDHERBYI (Fray et al. 1999) and contains NcoI and SphI sites overlapping the initiating ATG codon and the cleavage site of the encoded chloroplast transit peptide fragment, respectively.
Genes encoding proteins to be targeted to chloroplasts can be cloned into pBSRBCS using the SphI site for generating the ATG codon of the desired gene in frame with the RBCS sequence.
Plasmid pBC35dcm was designed to direct the constitutive expression of a plastid targeted E. coli Dcm methylase in planta. The dcm gene was PCR-amplified from the E. coli strain DH5α with oligonucleotides dcmF (TGCGCATGCAGGAAAATATATCAGTAAC) and dcmR (AGATCTAGATTATCGTGAACGTCGGCCATGTTG) introducing an SphI site overlapping the ATG codon at the 5′ end and a BamHI site at the 3′ end. The amplified fragment was cloned into the SphI and BamHI sites of the pBSRBCS. The RBCS-dcm cassette was excised as a KpnI/XbaI fragment and ligated to pBC35 cut with the same enzymes to give the plasmid pBC35dcm. pBC35 is a pBIN19 binary vector derivative (Bevan 1984), into which the Cauliflower Mosaic Virus (CaMV) 35S promoter, multiple cloning site and CaMV 35S terminator cassette PCR amplified from pDH51 (Pietrzak et al. 1986), was cloned between the EcoRI and HindIII sites (R.G. Fray, unpublished).
Binary vectors were transferred to Agrobacterium tumefaciens strain LBA4404 and used to transform sterile tobacco leaves as previously described (Fray et al. 1999).
DNA blot analysis
About 50 μg (or 1.5 μg for the analysis of chloroplast DNA) of total cellular DNA was digested with appropriate restriction enzymes, electrophoresed on 0.8% agarose gels and transferred to Hybond N+ (Amersham, UK) membranes using a standard neutral transfer procedure (Sambrook et al. 1989). DNA probes were random prime labelled with [α-32P] dCTP using a RediprimeTM II (Amersham, UK) kit. Probes were hybridised to the filters in: 10× Denhardt’s, 1% SDS, 5× SSPE, 1 mg ml−1 sonicated salmon sperm DNA at 65°C. Membrane washings were carried out with 2×, 1× and 0.1× SSPE, 0.1% SDS at 65°C, 10 min each. Signals were detected using auto-radiographic film (KODAK X-OMAT AR). All probes were cloned, fully sequenced and gel purified prior to use. Homoplastomy was confirmed using a 937 bp ndhF probe equivalent to fragment 114,316–115,253 of the chloroplast genome. The same probe was used to assess susceptibility to FokI digestion in the short single copy (SSC) sequence. For further cpDNA methylation analysis of FokI sites in the large single copy (LSC), a 1,268 bp fragment of the accD gene corresponding to nucleotides 59,057–60,325 of the chloroplast genome was used. To assay FokI sites in the inverted repeat sequences (IR), probe chIRE (nucleotides 95,327–96,322 IRB, 13,729–146,304 IRA) was used.
For analysis of sites affected by Dcm methylation, four fragments were generated to probe ScrFI cut cpDNA. Two fragments were used to assay ScrFI sites in the LSC–a 800 bp rbcL fragment (nucleotides 57,730–58,531) and a 1,066 bp chlA fragment (nucleotides 25,042–26,108 of the chloroplast genome). A 995 bp chlB probe (nucleotides 27,148–28,143) was used to assay ScrFI sites in theSSC and a 1,067 bp chlRD probe (nucleotides 93,351–94,418 IRB, 148,212–149,279 IRA) to assay ScrFI sites in the inverted repeats.
Results
Generation and analysis of transplastomic mFokI plants
Plastid transformation experiments with pFaadMet2 resulted in several spectinomycin resistant lines. After further rounds of selection, the homoplastomic state was confirmed for six of these by Southern analysis (Fig. 1b). MFokI methylates the sequence GGATG, which occurs 237 times in the plastid genome. To test for the presence of such methylation, total DNA from wild type and transplastomic plants was cut with BglII, an enzyme that recognizes a different target sequence and which is unaffected by FokI methylation, or with both BglII and FokI and the resulting fragments analysed by Southern blotting. Probes were chosen such that multiple FokI sites could be assayed in the small single copy (SSC), large single copy (LSC) and inverted repeat (IR) regions of the plastid genome.
Southern analysis using the ndhF probe adjacent to the transgene insertion site in the SSC, detects hybridizing fragments of 4,746 bp and 8,024 bp in the wild type and transplastomic plants, respectively following BglII digestion alone (Figs. 1b, 2a). There are four FokI sites within the wild type BglII fragment and a further two within the transgene sequence. Digestion with both BglII and FokI gives the expected hybridizing bands of 2,531 bp and 1,255 bp for wild type. However, in the case of the transplastomic plants, digestion with both enzymes gives a single band indistinguishable from that produced by a BglII digestion alone, indicating that all six FokI recognition sites are protected from FokI digestion in the transplastomic DNA (Fig. 2a). Using the accD probe, susceptibility to digestion of two FokI sites within the LSC was assayed. In both wild type and transplastomic plants, digestion with BglII results in a hybridizing fragment of 2,666 bp. Upon digestion with both BglII and FokI, the two predicted hybridizing bands of 1,287 bp and 578 bp are seen in the digestion of the wild type DNA, but for the transplastomic DNA, the hybridizing band remains 2,666 bp in size, indicating that both FokI sites are protected (Fig. 2b). Using probe chIRE from the IR, a single hybridizing band of 1,685 bp is obtained following BglII digestion of both wild type and transplastomic DNA. Following digestion with both BglII and FokI, additional cleavage at the three FokI sites should give hybridizing bands of 673 bp, 414 bp and 223 bp. In the wild type, the double digest gave a single band of 673 bp, consistent with complete digestion by these enzymes (the two smaller bands being run off the end of the gel) (Fig. 2c). When the transplastomic DNA is digested with both enzymes, bands of 1,685 bp (no FokI digestion) and 673 bp (full digestion) are seen. A number of additional digestion products are evident and their derivation can be fully explained as partial digestion products resulting from incomplete protection of the IR FokI sites (Fig. 2c).
Southern analysis indicates that FokI sites are protected from digestion to varying degrees in transplastomic plants. a Digestion with BglII gives ndhF hybridizing fragments of 4,746 bp and 8,024 bp for the wild type (WT) and transplastomic (T) plants respectively. Digestion with both BglII and FokI gives bands of 2,531 and 1,255 bp for wild type but protection of all six FokI sites in the transplastomic plant means that the 8,024 bp fragment is not digested further. b FokI sites within the LSC were assayed using the accD probe indicated. In both wild type and transplastomic plants, digestion with BglII gives a fragment of 2,666 bp. Digestion with both BglII and FokI, gives the two predicted bands of 1,287 and 578 bp for wild type, but protection of the two FokI sites leaves the 2,666 bp fragment unaltered in the transplastomic plant. c FokI sites within the IR were tested using the chIRE probe indicated. Digestion of both wild type and transplastomic cpDNA with BglII gives a single band of 1,685 bp. Digestion of wild type with both BglII and FokI gives a band of 673 (the smaller 414 bp and 223 bp fragments have been run out of the gel). Digestion of transplastomic DNA with both enzymes gives bands of 1,685 (no FokI digestion) and 673 bp (full digestion). A number of additional partial digestion products are evident and their derivation can be explained as follows; 1,310 bp (A + B + C), 1,271 bp (B + C + D), 1,048 bp (C + D), 896 bp (B + C) B BglII, F FokI, WT wild type, T transplastomic
Transplastomic plants expressing the mFokI methylase grew well with no obvious deleterious phenotypes observed as a result of MFokI induced adenosine cpDNA methylation. Reciprocal crosses were performed between transplastomic FaadMet2 and wild type plants and seeds were sown on MSR3 media supplemented with spectinomycin. 100% of germinating seeds were spectinomycin resistant when FaadMet2 was the maternal parent (Fig. 3a) and no spectinomycin resistant progeny were seen when it was the pollen donor (Fig. 3b). Two independent FaadMet2 lines were tested in this way and more than 10,000 seeds assayed in total. Complete methylation of FokI sites hybridizing to the SSC accD probe was confirmed for three of the spectinomycin resistant progeny (not shown).
The FaadMet2 transgene is maternally inherited. Reciprocal crosses were performed between transplastomic FaadMet2 and wild type plants and seedlings were screened for spectinomycin resistance. 100% of germinating seeds were spectinomycin resistant when FaadMet2 was the maternal parent (a) and no spectinomycin resistant progeny were seen when it was the pollen donor (b)
Generation and analysis of dcm transgenic plants
To test the effect of cytosine methylation, plants were transformed with a gene encoding the E. coli Dcm methylase equipped with a plastid import sequence. The dcm gene product methylates cytosines within the target sequence CCWGG (W = A or T). There are 171 such sites within the tobacco chloroplast genome. Transformed plants expressing the dcm methylase construct were analysed for the presence of cpDNA methylation by digestion with ScrFI. This enzyme cuts at the sequence CCNGG but is sensitive to cytosine methylation. Thus, in the presence of full Dcm methylation, CCAGG and CCTGG sites will become protected whilst CCCGG and CCGGG will remain susceptible to digestion. An rbcL probe (in the LSC) was chosen such that in the absence of methylation a fragment of 797 bp should result from digestion with ScrFI, whilst protection of a CCWGG site will result in a hybridising fragment of 1,810 bp (Fig. 4a). Extensive or complete methylation was seen at the assayed sites. A further eight sites were assayed in the LSC using chlA as probe. In the absence of Dcm methylation, digestion of wild type DNA with ScrF1 gives chlA hybridizing bands of 505, 461, 360 as well as a 27 bp fragment (not detected). Protection of all eight CCWGG sites in Dcm methylating lines should result in two hybridizing fragments of 874 and 2,738 bp. The presence of additional faint intermediate bands observed may indicate partial methylation of some DNA molecules (Fig. 4b). An additional eight CCWGG sites were assayed in a similar way in the SSC and IR using probes chlB and chlRD. Probe chlB (SSC) hybridizes to a single fragment of 1,082 bp following digestion with ScrFI when the two internal CCWGG sites are methylated. In the wild type plants, the absence of CCWGG methylation results in fragments of 731, 290 and 61 bp. Complete methylation of the two target sites was observed in transgenic plants (Fig. 4c). Probe chlRD (IR) was used to assess methylation within the IR sequences. In wild type plants, complete digestion with ScrFI gives hybridizing fragments of 424, 364, 317, 311 and 161 bp. Methylation of Dcm target sites should give two hybridizing bands of 583and 2,151 bp. The presence of additional intermediate bands may indicate partial methylation.
Southern analysis shows cpDNA from the transgenic dcm methylase plant is resistant to ScrFI cleavage at CCWGG sites. a In the absence of methylation, the rbcL probe (LSC) detects a fragment of 797 bp following digestion with ScrFI (WT lane). Protection of CCWGG sites gives a fragment of 1,810 bp (DCM). b Probe chlA was used to assay eight more CCWGG sites within the LSC. In the wild type, ScrFI digestion gives fragments of 505, 461, 360 and 27 bp (not visible). In the Dcm line, two fragments of 874 and 2,738 bp are generated as a result of protection of all eight CCWGG sites. The presence of additional intermediate bands may indicate partial methylation. c Probe chlB (SSC) hybridizes to a single 1,082 bp ScrFI fragment when the two internal CCWGG sites are methylated in the Dcm plant. In the wild type plants, the absence of CCWGG methylation results in fragments of 731, 290 and 61 bp. d Probe chlRD (IR) detects ScrFI fragments of 424, 364, 317, 311 and 161 bp in wild type cpDNA. In the Dcm plant, methylation of all six CCWGG sites results in two hybridizing bands of 583and 2,151 bp. The presence of additional intermediate bands may indicate partial methylation in some DNA molecules
No growth defects were seen as a result of this cytosine methylation.
Discussion
We have shown that bacterial restriction methylases can be expressed in the plastid environment and despite extensive adenosine or cytosine methylation of the chloroplast genome, plants appeared phenotypically normal. For an outbreeding suppression system based upon bacterial restriction/modification enzymes to work effectively, the entire target sites in the plastid genome should be protected from the endonuclease activity. In the mFokI transplastomic plants, apparently full target methylation was seen at eight sites within the assayed BglII fragments of SSC and LSC. However, incomplete methylation occurred at the three sites within the BglII fragment from the inverted repeats. This incomplete methylation in the IR may simply be a result of stocheometry (there are two copies of the IR sequence for every one copy of the LSC sequence), or it may be an indication that sequences within the IR are less accessible to the methylase–perhaps due to their relative proximity to the IR sequences involved in initiating DNA replication (Kunnimalaiyaan et al. 1997).
For the transgenic plants expressing a nuclear encoded, plastid targeted Dcm methylase, a total of 23 Dcm target sites were tested by ScrF1 digestion. The presence of the predicted higher molecular weight bands from the transgenic plants indicates that all the assayed sites are normally methylated. However, the feint bands of intermediate size seen when using the IR and one of the LSC probes may indicate that methylation is incomplete in some DNA molecules (Fig. 4).
For an out crossing inhibition scheme based upon bacterial restriction systems to work effectively, full methylation of all target sites will be required. Although extensive methylation was observed in both the FokI and Dcm plants, some sites were not fully protected. Thus, it may be necessary to test a number of alternative methylase and endonuclease pairs. There may be an optimum number of times that a target sequence should occur within the plastid genome and it may be best to avoid enzymes sites that are present within particular regions of the plastome (such as the IR). In addition it would be necessary to select enzymes that perform well under the salt, pH and temperature ranges found within the plastids of the target crop plant. The lethality of plastid-targeted restriction endonucleases needs to be tested and it may be necessary to avoid restriction enzymes with known off target “star” activity. However, the fact that both adenosine and cytosine methylation of the plastid genome appears to be readily tolerated means that there are potentially hundreds of methylase/endonucleas pairs that could be considered when constructing such an outcross prevention system.
References
Bevan M (1984) Binary Agrobacterium vectors for plant transformation. Nucl Acids Res 12:8711–8721
Bickle TA, Kruger DH (1993) Biology of DNA restriction. Microbiol Rev 57:434–450
Bock R (1998) Analysis of RNA editing in plastids. Methods 15:75–83
Burton WG, Grabowy CT, Sager R (1979) Role of methylation in the modification and restriction of chloroplast DNA in Chlamydomonas. Proc Nat Acad Sci USA 76:1390–1394
Dean C, van den Elzen P, Tamaki S, Black M, Dunsmuir P, Bedbrook J (1987) Molecular characterization of the rbcS multi-gene family of petunia (Mitchell). Mol Gen Genet 206:465–474
Eibl C, Zou ZR, Beck A, Kim M, Mullet J, Koop HU (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19:333–345
Fojtová M, Kovarík A, Matyásek R (2001) Cytosine methylation of plastid genome in higher plants. Fact or artefact? Plant Sci 160(4):585–593
Fray RG, Throup JP, Daykin M, Wallace A, Williams P, Stewart GSAB, Grierson D (1999) Plants genetically modified to produce N-acylhomoserine lactones communicate with bacteria. Nat Biotechnol 17(10):1017–1020
Gauly A, Kössel H (1989) Evidence for tissue-specific cytosine-methylation of plastid DNA from Zea mays. Curr Genet 15(5):371–376
Kita K, Kotani H, Sugisaki H, Takanami M (1989) The FokI restriction-modification system 1. Organization and nucleotide-sequences of the restriction and modification genes. J Biol Chem 264:5751–5756
Kobayashi H, Ngernprasirtsiri J, Akazawa T (1990) Transcriptional regulation and DNA methylation in plastids during transitional conversion of chloroplasts to chromoplasts. EMBO J 9(2):307–313
Kunnimalaiyaan M, Shi F, Nielsen BL (1997) Analysis of the tobacco chloroplast DNA replication origin (oriB) downstream of the 23 S rRNA gene. J Mol Biol 268(2):273–283
Marano MR, Carrillo N (1991) Chromoplast formation during tomato fruit ripening. No evidence for plastid DNA methylation. Plant mol biol 16(1):11–19
Marinus MG, Morris RN (1973) Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacreriol 114(3):1143–1150
Murray NE (2000) Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev 64:412–434
Ngernprasirtsiri J, Kobayashi H, Akazawa T (1988) DNA methylation as a mechanism of transcriptional regulation in nonphotosynthetic plastids in plant cells. Proc Natl Acad Sci USA 85(13):4750–4754
Nishiyama R, Wada Y, Mibu M, Yamaguchi Y, Shimogawara K, Sano H (2004) Role of a nonselective de novo DNA methyltransferase in maternal inheritance of chloroplast genes in the green alga Chlamydomonas reinhardtii. Genetics 168:809–816
Ohta N, Sato N, Kawano S, Kuroiwa T (1991) Methylation of DNA in the chloroplasts and amyloplasts of the pea, Pisum sativum. Plant Sci 78(1):33–42
Pietrzak M, Shillito RD, Hohn T, Potrykus I (1986) Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res 14:5857–5868
Ruf S, Karcher D, Bock R (2007) Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA 104:6998–7002
Sager R (1972) Cytoplasmic genes and organelles. Academic Press, New York
Sager R, Lane D (1972) Molecular basis of maternal inheritance. Proc Natl Acad Sci USA. 69:2410–2413
Sager R, Ramanis Z (1973) The mechanism of maternal inheritance in Chlamydomonas: biochemical and genetic studies. Theor Appl Genet 43(3–4):101–108
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sugisaki H, Kita K, Takanami M (1989) FokI restriction-modification system II. Presence of two domains in FokI methylase responsible for modification of different DNA strands. J Biol Chem 264:5757–5761
Svab Z, Maliga P (2007) Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA 104:7003–7008
Svab Z, Hajdukiewicz P, Maliga P (1990) Stable transformation of plastids in higher-plants. Proc Natl Acad Sci USA 87:8526–8530
Takio S, Satoh Y, Satoh T (1994) Occurrence of DNA methylation in chloroplasts of the suspension cultured cells from a liverwort, Marchantia paleacea var. diptera. J Plant Physiol 143(2):173–177
Umen JG, Goodenough UW (2001) Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev 15(19):2585–2597
Waters MT, Fray RG, Pyke KA (2004) Stromule formation is dependent upon plastid size, plastid differentiation status and the density of plastids within the cell. Plant J 39:655–667
Acknowledgments
This work was supported by a Biotechnology and Biological Sciences Research Council grant D17264 awarded to RGF which is gratefully acknowledged. We would like to thank Donald Grierson for helpful comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W. Harwood.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Jaffé, B., Kovács, K., Andras, C. et al. Methylation of chloroplast DNA does not affect viability and maternal inheritance in tobacco and may provide a strategy towards transgene containment. Plant Cell Rep 27, 1377–1384 (2008). https://doi.org/10.1007/s00299-008-0567-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0567-0