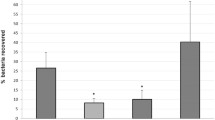
Abstract
Salmonella, a genus that is closely related to Escherichia coli, includes many pathogens of humans and other animals. A notable feature that distinguishes Salmonella from E. coli is lactose negativity, because the lac operon is lost in most Salmonella genomes. Here, we expressed the lac operon in Salmonella enterica serovar Typhimurium and compared the virulence of the Lac+ strain to that of the wild-type strain in a murine model, invasion assays, and macrophage replication assays. We showed that the Lac+ strain is attenuated in vivo and the attenuation of virulence is caused by its defect in epithelial cell invasion. However, the invasion-defective phenotype is unrelated to lactose utilization. Through sequencing and the comparison of the transcriptome profile between the Lac+ and wild-type strains during invasion, we found that most flagellar genes were markedly downregulated in the Lac+ strain, while other genes associated with invasion, such as the majority of genes encoded in Salmonella pathogenicity island 1, were not differentially expressed. Moreover, we discovered that lacA is the major repressor of flagellar gene expression in the lac operon. In conclusion, these data demonstrate that the lac operon decreases Salmonella invasion of epithelial cells through repression of flagellar biosynthesis. As the ability to invade epithelial cells is a critical virulence determinant of Salmonella, our results provide important evidence that the loss of the lac operon contributes to the evolution of Salmonella pathogenicity.




Similar content being viewed by others
References
Beltran P, Musser JM, Helmuth R, Farmer JR, Frerichs WM, Wachsmuth IK, Ferris K, McWhorter AC, Wells JG, Cravioto A, Et A (1988) Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A 85:7753–7757
Bera A, Biswas R, Herbert S, Gotz F (2006) The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun 74:4598–4604
Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B (2000) Salmonella nomenclature. J Clin Microbiol 38:2465–2467
Briones V, Tellez S, Goyache J, Ballesteros C, Del PLM, Dominguez L, Fernandez-Garayzabal JF (2004) Salmonella diversity associated with wild reptiles and amphibians in Spain. Environ Microbiol 6:868–871
Chattopadhyay S, Paul S, Kisiela DI, Linardopoulou EV, Sokurenko EV (2012) Convergent molecular evolution of genomic cores in Salmonella enterica and Escherichia coli. J Bacteriol 194:5002–5011
Coburn B, Grassl GA, Finlay BB (2007) Salmonella, the host and disease: a brief review. Immunol Cell Biol 85:112–118
Crosa JH, Brenner DJ, Ewing WH, Falkow S (1973) Molecular relationships among the Salmonelleae. J Bacteriol 115:307–315
de Jong HK, Parry CM, van der Poll T, Wiersinga WJ (2012) Host-pathogen interaction in invasive salmonellosis. PLoS Pathog 8:e1002933
Doolittle RF, Feng DF, Tsang S, Cho G, Little E (1996) Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271:470–477
Eswarappa SM, Karnam G, Nagarajan AG, Chakraborty S, Chakravortty D (2009) lac repressor is an antivirulence factor of Salmonella enterica: its role in the evolution of virulence in Salmonella. PLoS ONE 4:e5789
Finlay BB, Brumell JH (2000) Salmonella interactions with host cells: in vitro to in vivo. Philos Trans R Soc Lond B Biol Sci 355:623–631
Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, Connor TR, Seth-Smith H, Vernikos GS, Robinson KS, Sanders M, Petty NK, Kingsley RA, Baumler AJ, Nuccio SP, Contreras I, Santiviago CA, Maskell D, Barrow P, Humphrey T, Nastasi A, Roberts M, Frankel G, Parkhill J, Dougan G, Thomson NR (2011) Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog 7:e1002191
Giannella RA, Washington O, Gemski P, Formal SB (1973) Invasion of HeLa cells by Salmonella typhimurium: a model for study of invasiveness of Salmonella. J Infect Dis 128:69–75
Guise AJ, Budayeva HG, Diner BA, Cristea IM (2013) Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses 5:1607–1632
Hensel M (2004) Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol 294:95–102
Huang W, Wang Z, Lei QY (2014) Acetylation control of metabolic enzymes in cancer: an updated version. Acta Biochim Biophys Sin (Shanghai) 46:204–213
Ibarra JA, Steele-Mortimer O (2009) Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol 11:1579–1586
Isberg RR, Falkow S (1985) A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262–264
Jones BD, Lee CA, Falkow S (1992) Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun 60:2475–2480
Jones GW, Richardson LA, Uhlman D (1981) The invasion of HeLa cells by Salmonella typhimurium: reversible and irreversible bacterial attachment and the role of bacterial motility. J Gen Microbiol 127:351–360
Lan R, Reeves PR, Octavia S (2009) Population structure, origins and evolution of major Salmonella enterica clones. Infect Genet Evol 9:996–1005
Le Minor L, Veron M, Popoff M (1982) The taxonomy of Salmonella. Ann Microbiol (Paris) 133:223–243
Mahlknecht U, Hoelzer D (2000) Histone acetylation modifiers in the pathogenesis of malignant disease. Mol Med 6:623–644
Malik-Kale P, Jolly CE, Lathrop S, Winfree S, Luterbach C, Steele-Mortimer O (2011) Salmonella—at home in the host cell. Front Microbiol 2:125
Marcus SL, Brumell JH, Pfeifer CG, Finlay BB (2000) Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect 2:145–156
Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A (1998) “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci U S A 95:3943–3948
McClelland M, Florea L, Sanderson K, Clifton SW, Parkhill J, Churcher C, Dougan G, Wilson RK, Miller W (2000) Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res 28:4974–4986
Merhej V, Georgiades K, Raoult D (2013) Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors. Brief Funct Genomics 12:291–304
Mira A, Ochman H, Moran NA (2001) Deletional bias and the evolution of bacterial genomes. Trends Genet 17:589–596
Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, Yoshikawa M, Sasakawa C (1993) The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol Microbiol 9:459–468
Olsen JE, Hoegh-Andersen KH, Casadesus J, Rosenkranzt J, Chadfield MS, Thomsen LE (2013) The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol 13:67
Scheelings TF, Lightfoot D, Holz P (2011) Prevalence of Salmonella in Australian reptiles. J Wildl Dis 47:1–11
Schmidt H, Hensel M (2004) Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17:14–56
Schmitt CK, Ikeda JS, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, O’Brien AD (2001) Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect Immun 69:5619–5625
Schroeder GN, Hilbi H (2008) Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156
Sittka A, Pfeiffer V, Tedin K, Vogel J (2007) The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63:193–217
Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007
Zhang Y, Buchholz F, Muyrers JP, Stewart AF (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 20:123–128
Zhang Y, Muyrers JP, Testa G, Stewart AF (2000) DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol 18:1314–1317
Acknowledgments
This work was supported by the International Science & Technology Cooperation Program of China (2012DFG31680), National Key Program for Infectious Diseases of China (2013ZX10004216-001-001), and National Natural Science Foundation of China (NSFC) Program (31270003).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, L., Ni, Z., Wang, L. et al. Loss of the lac Operon Contributes to Salmonella Invasion of Epithelial Cells Through Derepression of Flagellar Synthesis. Curr Microbiol 70, 315–323 (2015). https://doi.org/10.1007/s00284-014-0720-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0720-7