Abstract
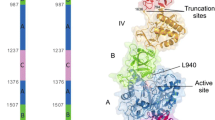
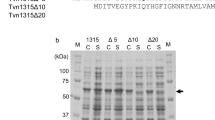
Clostridium sp. G0005 glucoamylase (CGA) is composed of a β-sandwich domain (BD), a linker, and a catalytic domain (CD). In the present study, CGA was expressed in Escherichia coli as inclusion bodies when the N-terminal region (39 amino acid residues) of the BD was truncated. To further elucidate the role of the N-terminal region of the BD, we constructed N-terminally truncated proteins (Δ19, Δ24, Δ29, and Δ34) and assessed their solubility and activity. Although all evaluated proteins were soluble, their hydrolytic activities toward maltotriose as a substrate varied: Δ19 and Δ24 were almost as active as CGA, but the activity of Δ29 was substantially lower, and Δ34 exhibited little hydrolytic activity. Subsequent truncation analysis of the N-terminal region sequence between residues 25 and 28 revealed that truncation of less than 26 residues did not affect CGA activity, whereas truncation of 26 or more residues resulted in a substantial loss of activity. Based on further site-directed mutagenesis and N-terminal sequence analysis, we concluded that the 26XaaXaaTrp28 sequence of CGA is important in exhibiting CGA activity. These results suggest that the N-terminal region of the BD in bacterial GAs may function not only in folding the protein into the correct structure but also in constructing a competent active site for catalyzing the hydrolytic reaction.







Similar content being viewed by others
References
Adam AC, Latorre-García L, Polaina J (2004) Structural analysis of glucoamylase encoded by the STA1 gene of Saccharomyces cerevisiae (var. diastaticus). Yeast 21:379–388. doi:10.1002/yea.1102
Aleshin AE, Feng PH, Honzatko RB, Reilly PJ (2003) Crystal structure and evolution of a prokaryotic glucoamylase. J Mol Biol 327:61–73. doi:10.1016/S0022-2836(03)00084-6
Bott R, Saldajeno M, Cuevas W, Ward D, Scheffers M, Aehle W, Karkehabadi S, Sandgren M, Hansson H (2008) Three-dimensional structure of an intact glycoside hydrolase family 15 glucoamylase from Hypocrea jecorina. Biochemistry 47:5746–5754. doi:10.1021/bi702413k
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 7:248–254
Carroll JD, Pastuszak I, Edavana VK, Pan YT, Elbein AD (2007) A novel trehalase from Mycobacterium smegmatis-purification, properties, requirements. FEBS J 274:1701–1714. doi:10.1111/j.1742-4658.2007.05715.x
Coutinho PM, Reilly PJ (1997) Glucoamylase structural, functional, and evolutionary relationships. Proteins 29:334–347. doi:10.1002/(SICI)1097-0134(199711)29:3<334::AID-PROT7>3.0.CO;2-A
Dalbøge H, Bayne S, Pedersen J (1990) In vivo processing of N-terminal methionine in E. coli. FEBS Lett 266:1–3. doi:10.1016/0014-5793(90)90001-B
Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S (1989) Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A 86:8247–8251
Hiromi K, Nitta Y, Numata C, Ono S (1973) Subsite affinities of glucoamylase: examination of the validity of the subsite theory. Biochim Biophys Acta 302:362–375
Kumar P, Satyanarayana T (2009) Microbial glucoamylases: characteristics and applications. Crit Rev Biotechnol 29:225–255. doi:10.1080/07388550903136076
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi:10.1038/227680a0
Li Z, Wei P, Cheng H, He P, Wang Q, Jiang N (2014) Functional role of β domain in the Thermoanaerobacter tengcongensis glucoamylase. Appl Microbiol Biotechnol 98:2091–2099. doi:10.1007/s00253-013-5051-2
Lin SC, Liu WT, Liu SH, Chou WI, Hsiung BK, Lin IP, Sheu CC, Dah-Tsyr Chang M (2007) Role of the linker region in the expression of Rhizopus oryzae glucoamylase. BMC Biochem 8:9. doi:10.1186/1471-2091-8-9
Marín-Navarro J, Polaina J (2011) Glucoamylases: structural and biotechnological aspects. Appl Microbiol Biotechnol 89:1267–1273. doi:10.1007/s00253-010-3034-0
Ohnishi H, Matsumoto H, Sakai H, Ohta T (1994) Functional roles of Trp337 and Glu632 in Clostridium glucoamylase, as determined by chemical modification, mutagenesis, and the stopped-flow method. J Biol Chem 269:3503–3510
Sakaguchi M, Matsushima Y, Nankumo T, Seino J, Miyakawa S, Honda S, Sugahara Y, Oyama F, Kawakita M (2014a) Glucoamylase of Caulobacter crescentus CB15: cloning and expression in Escherichia coli and functional identification. AMB Express 4:5. doi:10.1186/2191-0855-4-5
Sakaguchi M, Osaku K, Maejima S, Ohno N, Sugahara Y, Oyama F, Kawakita M (2014b) Highly conserved salt bridge stabilizes a proteinase K subfamily enzyme, Aqualysin I, from Thermus aquaticus YT-1. AMB Express 4:59. doi:10.1186/s13568-014-0059-2
Sakaguchi M, Shimodaira S, Ishida S, Amemiya M, Honda S, Sugahara Y, Oyama F, Kawakita M (2015) Identification of GH15 family thermophilic archaeal trehalases that function within a narrow acidic pH range. Appl Environ Microbiol 81:4920–4931. doi:10.1128/AEM.00956-15
Sambrook J, Russell DW (2012) Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Sevcík J, Solovicová A, Hostinová E, Gasperík J, Wilson KS, Dauter Z (1998) Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 Å resolution. Acta Crystallogr D Biol Crystallogr 54:854–866. doi:10.1107/S0907444998002005
Svensson B, Larsen K, Gunnarsson A (1986) Characterization of a glucoamylase G2 from Aspergillus niger. Eur J Biochem 154:497–502. doi:10.1111/j.1432-1033.1986.tb09425.x
Wilkinson AJ, Fersht AR, Blow DM, Winter G (1983) Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry 22:3581–3586
Acknowledgements
We are grateful to Yukari Saisaka (High-Tech Research Center, Meiji Pharmaceutical University) for performing the N-terminal sequence analysis and Syoma Sakamoto, Kazuaki Okawa, Satoshi Wakita, and Masahiro Kimura for their valuable suggestions and technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Funding
This study was supported in part by a grant from the Strategic Research Foundation Grant-aided Project for Private Universities of the Ministry of Education, Culture, Sport, Science, and Technology, Japan (MEXT) (S1411005); by the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan; and by the Project Research Grant from the Research Institute of Science and Technology, Kogakuin University.
Electronic supplementary material
ESM 1
(PDF 157 kb)
Rights and permissions
About this article
Cite this article
Sakaguchi, M., Matsushima, Y., Nagamine, Y. et al. Functional dissection of the N-terminal sequence of Clostridium sp. G0005 glucoamylase: identification of components critical for folding the catalytic domain and for constructing the active site structure. Appl Microbiol Biotechnol 101, 2415–2425 (2017). https://doi.org/10.1007/s00253-016-8024-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-8024-4