Abstract
The cruciferous fungal pathogen Verticillium longisporum represents an allodiploid hybrid with long spores and almost double the amount of nuclear DNA compared to other Verticillium species. V. longisporum evolved at least three times by hybridization. In Europe, virulent A1xD1 and avirulent A1xD3 hybrids were isolated from the oilseed crop Brassica napus. Parental A1 or D1 species are yet unknown whereas the D3 lineage represents Verticillium dahliae. Eleven V. longisporum isolates from Europe or California corresponding to hybrids A1xD1 or A1xD3 were compared. A single characteristic type of nuclear ribosomal DNA could be assigned to each hybrid lineage. The two avirulent A1xD3 isolates carried exclusively D3 ribosomal DNA (rDNA) which corresponds to V. dahliae. The rDNA of all nine A1xD1 isolates is identical but distinct from D3 and presumably originates from A1. Both hybrid lineages carry two distinct isogene pairs of four conserved regulatory genes corresponding to either A1 or D1/D3. D1 and D3 paralogues differ in several single nucleotide polymorphisms. Southern hybridization patterns confirmed differences between the A1 and D1/D3 isogenes and resulted in similar patterns for D1 and D3. Distinct signatures of the Verticillium transcription activator (VTA)2 regulatory isogene pair allow identification of V. longisporum hybrids by a single polymerase chain reaction and the separation from haploid species as V. dahliae or Verticillium albo-atrum. The combination between VTA2 signature and rDNA type identification represents an attractive diagnostic tool to discriminate allodiploid from haploid Verticillia and to distinguish between A1xD1 and A1xD3 hybrids which differ in their virulence towards B. napus.
Similar content being viewed by others
Introduction
Speciation is a dynamic process, which requires the isolation of different populations to prevent the exchange of genetic material. Reproductive barriers are formed by changes in ploidy, chromosomal rearrangements or incompatibilities between genes. Reproductive barriers are prerequisites for the evolution of diverging genetic elements, which convey an ecological separation (Mallet 2007; Giraud et al. 2010).
The fungal genus Verticillium has a world-wide distribution and includes species which cause vascular wilting disease and early senescence in a broad range of economically important crops including alfalfa, lettuce, hops, olive trees, oilseed rape, potato, tomato or strawberries (Pegg and Brady 2002). Verticillium morphology includes a characteristic verticilliate arrangement of the three to five asexual spore producing cells (phialides) forming branches at each node of the conidiophores (Kim et al. 2001). Verticillium dahliae and Verticillium albo-atrum are two closely related but distinct mature species. V. albo-atrum forms melanized resting mycelium, whereas the V. dahliae hyphae are not black. Instead, V. dahliae forms resting black microsclerotia, which are melanized clumps formed by budding of mycelial cells (Reinke and Berthold 1879; Isaac 1967; Goud et al. 2003). There is an overlap in host specificity, but both fungi can be distinguished by infecting different cultivars of a host plant species (Kim et al. 2001; Zeise and von Tiedemann 2002a). V. dahliae has a significantly broader host range and is able to infect more than 200 plant species. V. albo-atrum has in addition a specific host adaptation to lucerne and other leguminoses or to hops from the cannabaceae family (Pegg and Brady 2002; Agrios 2005; Vallad et al. 2005; Klosterman et al. 2009).
The list of hosts infected by Verticillium species is expanding and there is a continuous increase in severity of disease outbreaks on known hosts (Pegg and Brady 2002; Inderbitzin et al. 2011). Therefore, Verticillium represents an interesting and relevant model to study speciation by comparing nascent to already significantly diverged mature species. Crucifers are hardly infected by V. dahliae or V. albo-atrum. However, in 1961 a Verticillium was isolated from wilted horseradish as a first member of the crucifer family of Brassicaceae (Stark 1961). The isolated fungus forms preferentially only three phialides per node and survives by means of black microsclerotia which are elongated when compared to V. dahliae (Karapapa et al. 1997). The name Verticillium longisporum refers to twice (7.1–8.8 μm) as long asexual spores (conidia) in comparison to V. dahliae (3.5–5.5 μm; Karapapa et al. 1997; Zeise and von Tiedemann 2001; Collins et al. 2003). The numbers of V. longisporum isolates from crucifers are increasing and include horseradish from Illinois (Eastburn and Chang 1994), oilseed crops from Europe and Canada (Heale and Karapapa 1999; Zeise and von Tiedemann 2001; Steventon et al. 2002; Gladders et al. 2011) or cauliflower from California (Koike et al. 1994).
V. longisporum isolates are “near-diploid” or amphihaploid fungi with higher nuclear DNA amounts (about 1.8 times) than those of V. dahliae or V. albo-atrum isolates (Karapapa et al. 1997; Steventon et al. 2002; Collins et al. 2003). This almost diploid status might be the reason why numerous mutagenesis approaches have failed (Ingram 1968; Hastie 1973; Jackson and Heale 1985; Nagao et al. 1994; Subbarao et al. 1995; Zeise and von Tiedemann 2001). Most filamentous ascomycetes are haploid (Glass and Kuldau 1992). Experimental studies with the model fungus Aspergillus nidulans suggest that during adaptation to a novel environment, haploids deriving from diploids by parasexual recombination reach a higher fitness than the original diploids (Schoustra et al. 2007). Short-spored crucifer isolates might be haploid recombinants of long-spored isolates and V. longisporum distinct from noncrucifer isolates of V. dahliae (Karapapa et al. 1997; Barbara and Clewes 2003; Collins et al. 2003; Qin et al. 2006; Clewes et al. 2008).
Amplified fragment length polymorphism and restriction fragment length polymorphism originally suggested V. longisporum as interspecies hybrid between V. dahliae and V. albo-atrum or of close relatives of these two species (Karapapa et al. 1997; Steventon et al. 2002; Collins et al. 2003; Fahleson et al. 2004; Clewes et al. 2008; Collado-Romero et al. 2010). A recent comparison of eight genetic loci including the ribosomal internal transcribed spacer (ITS) regions and genes for two structural proteins (actin, tubulin), one transporter (mitochondrial oxalacetate transport protein), one translation factor (EF1-α), two enzymes (glyceraldehyde-3-P-dehydrogenase, tryptophan synthase) and the genes for the mating types (MAT1-1) suggested that V. longisporum hybridized at least three times resulting in the lineages A1xD1, A1xD2, A1xD3, with A1xD2 exclusively found in the USA. The A1 and D1 had been described as yet unknown Verticillium species, whereas D2 and D3 represent V. dahliae lineages (Inderbitzin et al. 2011).
In this work, we compared V. longisporum A1xD1 hybrids from Europe and California which are virulent for oilseed rape and cauliflower, respectively, with avirulent A1xD3 isolates from European oilseed rape. We focused on rDNA and four regulatory gene pairs to identify molecular differences on a genomic level which allow an accurate molecular diagnosis to discriminate allodiploid hybrids from each other and from haploid Verticillium relatives.
Materials and methods
Fungal isolates and maintenance
Twenty-six isolates of Verticillium species from different hosts were used in this study (Table 1). For the long-term maintenance, the isolates were grown separately in liquid Czapek-Dox or simulated xylem medium (SXM; Neumann and Dobinson 2003) for 10 days at 25 °C. Conidia were harvested by filtering the culture through Miracloth (Calbiochem, Darmstadt, Germany) the filtrate was centrifuged at 5,000 rpm for 10 min. After a washing step with sterile water, the pellet was resuspended in sterile water and the number of spores was counted under a microscope. Glycerin was added to the spore suspension at the final concentration of 20 % and aliquots of the spore suspension were frozen in liquid nitrogen and stored at −80 °C.
Isolation of nucleic acids
The isolates were grown for one week, at 25 °C in potato dextrose broth (Sigma-Aldrich Chemie GmbH, Munich, Germany) for genomic DNA or liquid SXM (Neumann and Dobinson 2003) for total RNA extraction. The fungal mycelia harvested with Miracloth (Calbiochem, Darmstadt, Germany) were ground to fine powder in liquid nitrogen. The fungal powder was used directly for nucleic acid extraction or stored at −80 °C for later use. Genomic DNA was extracted from the fungal powder (Kolar et al. 1988). Total RNA was extracted by using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total cDNA was generated from the total RNA by using RevertAid First-Strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany).
PCR amplification, cloning and sequencing
Polymerase chain reaction (PCR) amplifications were performed in 25 μl volumes with the high-fidelity Phusion polymerase (Finnzymes, Finland) using the following general conditions: an initial denaturation at 98 °C for 3 min; a cycle including denaturation at 98 °C for 30 s, annealing at 55–60 °C for 40 s, and extension at 72 °C for 1–4.5 min was repeated for 25–30 times; a final extension at 72 °C for 7 min and stored at 4 °C until used. PCR products were analyzed on a 1 % agarose gel and the DNA fragments were excised and purified with QIAquick gel extraction kit (Qiagen, Hilden, Germany). The purified DNA fragments were used for direct sequencing. For cloning, each DNA fragment of interest was amplified from V. longisporum genomic DNA by the high-fidelity Phusion polymerase with modifications optimized for interspecies hybrid microorganisms including increases in concentration of each primer (100 pmol) and each dNTP (20 mM) for each reaction of 25 μl, an increase in time for elongation step (4.5 min) together with a decrease in number of PCR cycles (25 cycles; Beser et al. 2007; Inderbitzin et al. 2011). The PCR product was ligated into the pJET1.2/blunt cloning vector using the CloneJET™ PCR Cloning Kit (Fermentas, St. Leon-Rot, Germany). The ligation mixture was transformed into E. coli DH5α competent cells. Colony PCR with Hot-Star Taq MasterMix (Qiagen, Hilden, Germany) was used to screen positive colonies from each cloning procedure. At least, 10–15 positive clones were selected and grown in LB (Luria-Bertani) liquid medium containing 100 μg/ml of ampicillin. Recombinant plasmids were purified with the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany) for sequencing. The sequencing was performed with the primers (forward and reverse) from the cloning kit by the in-house Göttingen Genomics Laboratory. DNA alignments with the Geneious Pro v5.4 software (Drummond et al. 2011) were performed to detect artificially chimeric sequences generated by PCR as by-products for elimination.
Southern hybridization
The target genes contain no restriction sites for the enzymes used for genomic DNA digestion. A fragment (about 600 bp) of the target gene or the whole gene was amplified and labeled as probe for Southern hybridization. In brief, 20 μg of genomic DNA were digested with 3 μl of an appropriate restriction enzyme for overnight. The digested mixture was analyzed on a 1 % agarose gel, and DNA was transferred to an Amersham Hybond-N membrane (GE Healthcare, Munich, Germany) by blotting. The DNA on the membrane was overnight hybridized to a specific probe and the Amersham CDP-Star Detection reagent (GE Healthcare, Munich, Germany) was used to detect chemiluminescence signals according to the manufacturer’s instructions.
DNA analysis
Characterization of 18S rRNA gene, ITS1, 5.8S rRNA, ITS2, 28S rRNA, and intergenic region (IGS) of the rDNA was based on Genbank accession number AF104926 (Pramateftaki et al. 2000). For the other conserved genes, the introns and exons were determined by comparing the cDNA sequences with their genomic DNA sequences using the ClustalW (Thompson et al. 2002). Detection of repeated signatures in the DNA sequences was carried out with Repeats Finder implemented in the multiplatform open-source software Unipro UGENE v1.10 (Okonechnikov et al. 2012). For DNA analysis and comparison in more details, the commercial software Geneious Pro v5.4 (Drummond et al. 2011) was used. Phylogenetic trees were constructed with the MEGA 5.0 software (Tamura et al. 2011) based on the neighbor-joining method (Saitou and Nei 1987). Statistical reliabilities of the internal branches were assessed for the trees with bootstraps of 1,000 replicates. The p distance model was used and gaps/missing data were treated with complete deletion or pairwise deletion.
EMBL accessions
DNA sequences were deposited in the EMBL-Bank with the accession numbers HE972012–HE972156.
Results
rDNA of V. longisporum isolates is homogenized to D3 rDNA for avirulent and A1 rDNA for virulent lineages
Allodiploid V. longisporum species were suggested to be interspecific hybrids (Karapapa et al. 1997; Steventon et al. 2002; Collins et al. 2003; Fahleson et al. 2004; Clewes et al. 2008; Collado-Romero et al. 2010). We have analyzed and compared the rDNA from 24 Verticillium isolates of three species including 11 V. longisporum, 9 V. dahliae, and 4 V. albo-atrum that originated from different hosts of different geographical regions (Table 1) for a more comprehensive picture of the evolution of V. longisporum (Fig. 1). Parts of rDNA are commonly used as taxonomic markers (Schoch et al. 2012). The copy numbers of the individual rDNAs vary between 30 and 30,000 copies in eukaryotes (Rooney and Ward 2005; Ganley and Kobayashi 2007).
Analysis of ITS region of ribosomal DNA in Verticillium plant pathogens. a V. longisporum isolates carry only one type of ITS. The V. dahliae D3 type corresponds to nine V. dahliae isolates. The A1 type corresponding to the unknown species A1 is closer related to V. albo-atrum than to V. dahliae. The two pyrimidine exchanges between ITS1 of A1 and V. albo-atrum at positions 90 and 125 are indicated in bold. The ITS region comprises ITS1 (129 nucleotides), 5.8S rRNA and ITS2 (165 nucleotides). A1xD3 strains (Vl19 and Vl32) isolated from oilseed rape in Northern Germany are avirulent, whereas the A1xD1 strains from oilseed rape in Europe or from cauliflower in California are virulent. b ITS-based phylogenetic analysis. Avirulent A1xD3 isolates carry V. dahliae ITS, whereas virulent A1xD1 isolates carry A1 ITS closer related to V. albo-atrum strains. The tree includes two additionally analyzed Verticillia for comparison: V. tricorpus is distant from V. longisporum, V. dahliae, and V. albo-atrum; V. nigrescens Vni1 (now Gibellulopsis nigrescens) represents the root of the phylogenetic tree
Ribosomal DNA repeats including 18S rRNA gene, two internal transcribed spacers (ITS1, ITS2) covering 5.8S rRNA gene and a large IGS were analyzed (Figs. 1 and 2). PCR amplification using various primer pairs ITS-F/ITS-R and IGS-F/IGS-R (Table 2) resulted for each primer pair in only one single sequence.
Analysis of the rDNA IGS located between 28S rRNA and 18S rRNA gene of Verticillium isolates. a IGS sequences of indicated strains which vary in their sequences between 1.7 and 1.9 kb (Fig. S1) were compared by a phylogenetic tree. The IGS sequence of V. tricorpus distant from all three species is used as root. A1 IGS rDNA (from A1xD1) group is closer to V. albo-atrum rDNA, whereas the D3 IGS (from A1xD3) is the closest to V. dahliae Vd39 isolated from sunflower. b A1 18S rRNA length of virulent A1xD1 isolates was compared to the 18S rRNA of the other strains. A1 18S rRNA carries an additional intron of 839 nucleotides which is as well absent in 18S rRNA of D3 of avirulent A1xD3 isolates as in 18S rRNA of V. dahliae or V. albo-atrum isolates
The ITS-F and ITS-R primers amplified DNA includes a part of the 18S rRNA gene (212 bp), ITS1 sequence (129 bp), 5.8S rRNA gene (158 bp), ITS2 sequence (165 bp), and a part of 28S rRNA (178 bp). Sequence analyses revealed that the 5.8S rRNA gene located between ITS1 and ITS2 is completely conserved within all three species. All nine V. dahliae isolates are separated from all four V. albo-atrum isolates in the ITS1 and the ITS2 region by characteristic nucleotide polymorphisms (Fig. 1a).
There are two different ITS types (ITS type 1 and ITS type 2) within the 11 V. longisporum isolates. The ITS type 1 of the two European isolates Vl19 and Vl32 is identical to the ITS of V. dahliae. Vl19 and Vl32 have been isolated from oilseed rape in Europe and are both avirulent (Zeise and von Tiedemann 2002a, b). European V. longisporum isolates represent either the A1xD1 or A1xD3 hybrid lineages with D3 being the only parental strain which corresponds to V. dahliae (Inderbitzin et al. 2011). This suggests that Vl19 and Vl32 represent A1xD3 hybrids. The ITS type 2 is close to the ITS of V. albo-atrum except for two pyrimidine exchanges in the ITS1 (Fig. 1a). The ITS type 2 was detected for six virulent European isolates (Vl18, Vl40, Vl43, Vl82, Vl83, Vl84) from oilseed rape (Zeise and von Tiedemann 2002a) and three American virulent cauliflower isolates (Vl59, Vl60, Bob70; Zeise and von Tiedemann 2002a, b; Qin et al. 2006). A1 and D1 are unknown species which are distinct from V. dahliae or from V. albo-atrum. A1 has been described to be closer related to V. albo-atrum than to V. dahliae, which is supported by the phylogenetic tree analysis (Fig. 1b). Therefore the ITS type 2 was assigned as the ITS of the unknown V. albo-atrum related species A1.
A closer inspection of the IGS separating rDNA repeats revealed PCR products ranging from 1.7 to 1.9 kb using the primer pair IGS-F/IGS-R. The IGS region is highly polymorphic within all three species and can be divided into two parts based on the sequence conservation (Fig. S1). The variable part of approximately 1 kb starts at the end of the 28S rRNA gene and contains sequences which can be repeated and numerous single nucleotide polymorphisms. In contrast, the conserved part of about 0.7–0.9 kb is situated adjacent to the 18S rRNA gene and reveals high similarity within all three species. All results are summarized in a phylogenetic tree (Fig. 2a).
In agreement to the data obtained with the ITS, there are two different IGS sequence types (IGS types 1 and 2) within the 11 V. longisporum isolates. The IGS type 1 (two European isolates Vl19 and Vl32) groups within the V. dahliae clade and corresponds to D3 (Fig. 2a). It contains a single signature of 81 nucleotides (AGCTACCCGGGAATTGGACCAGTTTTGAGGCGGCAGCTACCCGGGAGTTGGCGAAAAACGACCAAGTCGGACACCTTGG). The single copy of the 81-nucleotide signature of the D3 IGS of A1xD3 hybrids corresponds to the IGS of V. dahliae Vd39 from sunflower, whereas there are two or three repeats of this sequence in the IGS of the other analyzed V. dahliae isolates (Fig. S1).
The IGS type 2 of the A1xD1 isolates is closer to the clade of the IGS region of V. albo-atrum and corresponds to A1 (Fig. 2a). The A1 IGS of the virulent V. longisporum A1xD1 lineages includes three copies of a 39 nucleotide repeat (ATCTGGGAGCTACCCGGGAGTTGGA AATTTGGAGAACGG) in the IGS region from European oilseed rape and four repeat copies of the same sequence in the IGS of the American A1xD1 isolates from cauliflower. V. albo-atrum IGS differs from A1 IGS and has no specific repeats in the IGS region of rDNA (Fig. S1).
We aimed to discriminate between the two V. longisporum hybrids A1xD1 (virulent) and A1xD3 (avirulent) by a single PCR reaction. The 18S rRNA gene-specific primer pair (18S-rRNA1/18S-rRNA2) resulted in a 0.44–1.28 kb fragment corresponding to two types of 18S rRNA genes for different V. longisporum isolates (Fig. 2b). Both types differ in an intron of 839 nucleotides, which is present in all 9 A1 rDNA V. longisporum isolates (Vl18, Vl40, Vl43, Vl59, Vl60, Bob70, Vl82, Vl83, Vl84). The intron is absent in the 18S rRNA genes of the two A1xD3 V. longisporum isolates (Vl19, Vl32) carrying the V. dahliae D3 rDNA type and is also absent in all analyzed V. dahliae and V. albo-atrum isolates. The 839-bp intron is therefore typical for the A1 lineage but only conserved within the virulent V. longisporum A1xD1 hybrids which have lost the D1 rDNA (Fig. 2b). A1 might have acquired this intron by horizontal gene transfer provided by another organism like a host plant (Karapapa and Typas 2001).
Our results demonstrate that the analyzed V. longisporum hybrids A1xD3 and A1xD1 which could be isolated from the same geographical region in northern Germany (Mecklenburg–Vorpommern) can be separated by their rDNA types. Only the two rDNA types A1 and D3 were maintained, the D1 rDNA type has been lost in all isolates. The virulent A1xD1 hybrids carried only the A1 rDNA including a specific intron of unknown origin which can be used to discriminate these strains from the avirulent A1xD3 hybrids carrying exclusively the D3 rDNA type which resembles the rDNA of the V. dahliae isolates.
Both rDNA types of V. longisporum carry a pair of isogenes for putative regulators, which are conserved in ascomycetes
The molecular analysis of single nuclear genes is yet limited in V. longisporum and there is hardly any comparison of regulatory genes between the different Verticillium isolates corresponding to A1xD1, A1xD3 hybrids and related haploid species. Two orthologues of the velvet genes VeA and VelB were examined, which are conserved in ascomycetes. For several fungi, these regulators are known to control secondary metabolism and fungal development including hyphal morphogenesis or the formation of the conidiophore producing asexual spores (Bayram et al. 2008; Calvo 2008; Bayram and Braus 2012).
The VEL1-F/VEL1-R primers allowed only the amplification of a 1.718-kb fragment of VeA orthologues (VEL1) from genomic DNA of all nine V. dahliae isolates as well as from all 11 V. longisporum isolates (A1xD1 and A1xD3), but not from the four V. albo-atrum isolates. DNA sequencing resulted in the internal primer set VEL1-F/VEL1-612-R which amplified a 612-bp fragment of all isolates. The 612-bp products were cloned and sequenced and Fig. S2a summarizes the data in a phylogenetic tree. V. longisporum A1xD1 and A1xD3 carry pairs of isogenes for VEL1 (VlVEL1-1, VlVEL1-2). The VlVEL1-1 sequence of A1xD3 lineage (Vl19, Vl32) corresponds to parental D3 and is identical to V. dahliae VdVEL1. The D1 derivative VlVEL1-1 of A1xD1 hybrids (Vl40, Vl43, Bob70) has accumulated three nucleotide polymorphisms (SNPs). Both lineages of V. longisporum share the same VlVEL1-2 sequence which corresponds to the A1 parent and displays a 95 % identity to the single genes of V. dahliae VdVEL1 or V. albo-atrum VaVEL1 (Fig. S2a).
Southern hybridization verified the two paralogues of V. longisporum and allowed a comparison of the environment of the two gene loci within the genome with the related haploid strains of V. dahliae and V. albo-atrum (Fig. 3a). The Southern hybridization pattern of isolates representing both hybrids (A1xD1 and A1xD3) with the different rDNA types were indistinguishable and confirmed the presence of two isogene copies in contrast to the single copies of V. albo-atrum or V. dahliae.
Comparative Southern hybridization for four single genes of V. dahliae (Vd) and V. albo-atrum (Va) in comparison to two isogenes of two V. longisporum (Vl) strains with different rDNA. a Genomic DNA of all representative strains was digested with NheI and probed with labeled VEL1 DNA. The same genomic DNAs were also treated with PvuI and hybridized to a VEL2 probe (b), digested with NcoI with VTA1 probe (c), and digested with BglII and probed with labeled VTA2 (d)
Amplification of the VelB homologues using the primers VEL2-F/VEL2-R (Table 2) resulted in 1.615-1.618-kb fragments. Sequencing revealed two sequences (VlVEL2-1, VlVEL2-2) for VEL2 in V. longisporum isolates but only one in V. dahliae (VdVEL2) and V. albo-atrum (VaVEL2). The data are summarized in the phylogenetic tree of the Fig. S2b. VlVEL2-1 of A1xD3 corresponds to the D3 parent and is identical to V. dahliae VEL1 (VdVEL1). The corresponding D1 VlVEL2-1 allele of A1xD1 carries differences in some single nucleotides. Interestingly, the VEL2 sequences of V. dahliae Vd73 (from linseed) can be separated from other V. dahliae strains including the D3 parent of V. longisporum for VEL2 but not for VEL1. Similarly, V. dahliae isolate Vd14.01 (from pistachio) has accumulated even more SNPs which separates it even more from the other V. dahliae strains than the D1 parent of the V. longisporum A1xD1 hybrid for VEL2 but not for VEL1 (Fig. S2b). These SNP accumulations for VEL2 might reflect a specific yet unknown function of the VEL2 encoded regulator for host adaptation. The other V. longisporum VlVEL2-2 isoallele corresponds to A1 and is identical in both lineages of V. longisporum with an identity of about 95 % to both V. dahliae and V. albo-atrum genes.
These data suggest that similarly to VEL1, there are also two isogenes for VEL2 in the V. longisporum genome. Comparison of the Southern hybridization pattern of both rDNA types revealed two loci for VEL2 in V. longisporum including one locus which is similar to V. dahliae. The other Southern band is identical in both V. longisporum rDNA types but distinct to both V. dahliae and V. albo-atrum and presumably reflects the second parent A1 as unknown species which is grouped closer to V. albo-atrum than to the other parental strain (Fig. 3b). This further corroborates one common ancestor parent for both types of rDNA of V. longisporum.
Conclusively, our data demonstrate that in contrast to the rDNA repetitive genes, which are uniform, there are two isogenes for the putative regulators VEL1 and VEL2 in both V. longisporum lineages (A1xD1, A1xD3) with high identities (94–100 %) to each other and to the corresponding single genes of V. dahliae or V. albo-atrum as relatives to D1, D3, or A1. The reason why V. dahliae strains from specific hosts have accumulated more mutations in the VEL2 gene remains to be elucidated.
Paralogue transcription factor isogene pairs of V. longisporum
We extended our analysis by examining two orthologues of transcriptional regulatory genes, which are expressed in V. longisporum. Five thousand expressed sequence tags were sequenced from a cDNA library of V. longisporum (Singh et al. 2010). This resulted in the identification of transcripts for genes for the putative transcription factors Verticillium transcription activator (VTA)1 and VTA2. The corresponding genes encoding zinc finger family proteins which are conserved within filamentous ascomycetes (Fig. S3) were cloned and sequenced.
The VTA1 gene encodes a protein containing the Zn(II)2Cys6 conserved domain. This domain is similar to the conserved domain of the AflR transcription factor in Aspergilli required for mycotoxin biosynthesis of the cancerogenic aflatoxin (Yu et al. 1996). The regulator Fsr6 in Fusarium fujikuroi, an orthologue of Vta1 (Fig. S3a), has been recently described for its function in regulation of pigment biosynthesis (Studt et al. 2012). V. longisporum carries two isogenes for VTA1 which are both transcribed resulting in expressed sequence tags without introns. They correspond to single genes in V. dahliae or V. albo-atrum. The two VTA1 sequences of 1.275 and 1.278 kb display a 95–99.9 % identity to each other and to the corresponding genes of the haploid Verticillia with a significant different signature: a three-nucleotide sequence in the longer VTA1 sequence of all V. longisporum isolates (VlVTA1-1) is only present in V. dahliae. These VlVTA1-1 sequences of the two V. longisporum hybrids A1xD3 (Vl19, Vl32) and A1xD1 (Vl40, Vl43, Bob70) are almost identical to V. dahliae VdVTA1 with differences in one (D3) or eight (D1) single nucleotide substitutions of the 1.278-bp sequences, respectively. The V. albo-atrum as well as all shorter A1 VTA1 sequences of V. longisporum (VlVTA1-2) are identical in both lineages A1xD1 and A1xD3 and include the small three-nucleotide deletion in comparison to the isoallele (Figs. 4 and S4). Consistently, comparison of the Southern hybridization pattern revealed two loci for the VTA1 genes in both rDNA types of V. longisporum, where one locus results in the same pattern as V. dahliae, whereas there is a different pattern for the A1-derived gene which carries the same deletion as the V. albo-atrum gene (Fig. 3c). This further supports that A1 as the second parent species separates from its relative V. albo-atrum by several additional single nucleotide polymorphisms.
Two VTA1 isogenes of V. longisporum (VTA1-1, VTA1-2) in comparison to the corresponding single genes of V. dahliae and V. albo-atrum. a Comparative scheme of VTA1 from all three Verticillium species: Both the virulent A1xD1 isolates (Vl40, Vl43, Bob70) and the avirulent A1xD3 (Vl19, Vl32) carry two isogenes for VTA1 gene (VlVTA1-1, VlVTA1-2), whereas only one sequence of this gene is present in V. dahliae (VdVTA1) and V. albo-atrum (VaVTA1). The sequences for VlVTA1-1 of A1xD3 and A1xD1 are almost identical to V. dahliae VdVTA1 with a minor difference in one or eight SNPs, respectively. Both VlVTA1-1 and VdVTA1 possess the same signature consisting of a three-nucleotide insertion (TTA) in the coding region. In contrast, VlVTA1-2 is identical in both A1xD1 and A1xD3 and lacks the insertion of three nucleotides like V. albo-atrum VaVTA1. VlVTA1-2 shows 95–96 % identitiy to VdVTA2 and VaVTA2. Colored lines within the genes correspond to corresponding SNPs. Additional SNPs present in genes of the lower in comparison to the upper box are summarized next to the boxes and visualized by grey color. The VdVTA1 (from Vd73 isolate) and VaVTA1 (from Va1 isolate) are used as putative parent sequences for SNPs calculation. b Phylogenetic analysis based on the conserved VTA1 gene shows close connection of V. longisporum hybrids to V. dahliae and V. albo-atrum
The VTA2 gene containing a C2H2 conserved motif is an orthologue of the CON7 gene in the rice blast fungus Magnaporthe grisea. The Con7 protein controls approximately 100 genes and is essential for appressorium formation and pathogenicity (Odenbach et al. 2007). VTA2 genes are broadly conserved within filamentous fungi and the Verticillium isogenes are related to fungal plant pathogens such as Fusarium species and M. grisea (Fig. S3b). Comparison of the expressed sequence tags to the genomic sequences amplified by PCR revealed a complex gene structure for VTA2, which includes four introns and five exons. The V. dahliae and V. albo-atrum VTA2 orthologues vary slightly in size with 1.701 kb and 1.754–1.759, respectively. The first exon of V. dahliae VTA2 has an insertion of nine nucleotides (ACGCTCACC) when compared with V. albo-atrum VTA2. In addition, the third intron (I3) of the V. dahliae gene (217 bp) is 63–68 nucleotides shorter than V. albo-atrum I3 (280–285 bp). In contrast to the single parental genes, both types of V. longisporum rDNA carry two paralogues (Figs. 5 and S5).
The evolutionary origin of all 11 V. longisporum strains based on the nuclear gene VTA2 with the length of 1,701–1,759 nucleotides. a Comparative scheme of VTA2 from all three Verticillium species: VlVTA2-1 and VlVTA2-2 isogenes of both V. longisporum lineages A1xD1 and A1xD3 are derivatives from V. dahliae and V. albo-atrum, respectively. Characteristic signatures include a nine-nucleotide insertion (ACGCTCACC) in the first exon (E1) and the length of the third intron (I3). VlVTA2-1 and the V. dahliae VdVTA2 carries the nine-nucleotide insertion in the first exon and a shortened third intron (217 bp), whereas VlVTA2-2 and V. albo-atrum VaVTA2 lacks this insertion in the first exon and carries an extended third intron (280–285 bp). VlVTA2-1 of the avirulent A1xD3 isolates is identical to V. dahliae VdVTA2 and differs from VlVTA2-1 of the virulent A1xD1 isolates by 6 single nucleotide polymorphisms (SNPs). VlVTA2-2 is identical in all A1xD1 isolates but differs from VlVTA2-2 of A1xD3 by only a single nucleotide. VlVTA2-2 displays 97 % identity to V. albo-atrum VaVTA2 and only 93 % to V. dahliae VdVTA2. SNPs are indicated by colors. The VdVTA2 (from Vd73 isolate) and VaVTA2 (from Va1 isolate) are used as putative parent sequences for SNPs calculation. b Phylogenetic analysis for all 11 V. longisporum isolates in comparison to V. dahliae and V. albo-atrum isolates. The VTA2 sequence of V. tricorpus is used as root of the phylogenetic tree
The two VTA2 isogenes for V. longisporum (VlVTA2-1, VlVTA2-2) varied in lengths slightly between 1.701 and 1.755 kb. The VlVTA2-1 sequences from the two V. longisporum lineages A1xD1 and A1xD3 carry the same characteristic ACTCTCACC insertion in the first exon and the shortened third intron of 217 bp as V. dahliae VTA2 gene. The V. dahliae VTA2 gene is absolutely conserved in this species. VlVTA2-1 of two V. longisporum A1xD3 isolates (Vl19, Vl32) carrying V. dahliae rDNA corresponds to D3 because it is identical to VdVTA2. This allele differs from the D1 allele of VlVTA2-1 of the A1xD1 isolates carrying V. albo-atrum-related rDNA by six single nucleotides. In contrast, the A1 allele VlVTA2-2 of the A1xD1 lineage including six European isolates (Vl18, Vl40, Vl43, Vl82, Vl83, Vl84) and three American isolates (Vl59, Vl60, Bob70) is identical and differs from the other A1 allele of VlVTA2-2 of the A1xD3 lineage (Vl19, Vl32) by only a single nucleotide. This VlVTA2-2 sequence exhibits an identity of about 97 % to V. albo-atrum VaVTA2 gene. It carries the characteristic first exon of V. albo-atrum lacking the nine nucleotide insertion as well as the longer third intron of 282 bp (Figs. 5 and S5). Southern hybridization confirmed that the two V. longisporum VTA2 isogenes are either closer related to V. dahliae or to V. albo-atrum, respectively (Fig. 3d).
The VTA2 gene analyses corroborated that the two isogenes VlVTA2-1 and VlVTA2-2 are completely conserved within all nine V. longisporum A1xD1 isolates derived from Europe and America and have not (yet) been changed during geographical separation. Sequencing of the corresponding VTA2 genes from nine V. dahliae isolates from different geographical regions also reveals an absolute conservation (100 % identity) of this gene. In contrast, V. albo-atrum isolates display minor changes in VTA2 genes depending on hosts. The VTA2 genes from the non-Alfalfa isolates (Va1, Va3, Va4) deriving from potato, hop, and tomato are completely identical to each other, but exhibit a slight reduction of 97.72 % identity compared to VTA2 sequence of the Alfalfa isolate Va2. However, the Alfalfa and the non-Alfalfa group still have conserved the signatures of the first exon and the third intron (Figs. 5 and S5). This variation in nucleotide polymorphisms in V. albo-atrum might be the reason for the differences between the present V. albo-atrum isolates and the unknown V. albo-atrum related parental species A1 of the hybrid V. longisporum.
VTA2 is a barcode marker for V. longisporum
ITS region of nuclear rDNA as a commonly used universal DNA barcode marker for fungal taxonomy (Schoch et al. 2012) are useful to discriminate between V. dahliae and the closely related species V. albo-atrum. This method is limited for V. longisporum. As shown above, European V. longisporum hybrids carry either a single rDNA which corresponds to D3 V. dahliae rDNA or A1 rDNA which is closer to V. albo-atrum than to V. dahliae (Fig. 1). Hybrids with the D1 rDNA have not yet been described.
We searched for a simple molecular marker to discriminate between haploid and hybrid Verticillia by polymerase chain reaction. We initially tested two primer pairs for glyceraldehyde-3-P-dehydrogenase and mitochondrial oxalacetate transport protein genes that were reported to be specific for separation between A1xD1 and A1xD3 lineages (Inderbitzin et al. 2011). Using our strain collection, we found that both of these primer pairs do not always separate A1xD3 isolates from V. dahliae, although it is required to perform three different PCR reactions for each primer pair (Fig. S6). Furthermore, these primers were designed on basis of differences in some single nucleotide polymorphisms between two isogenes in the hybrids; therefore optimized PCR procedures are required to avoid unspecific amplifications of the homologous DNA fragments.
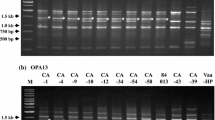
We therefore tested whether the distinct VTA2 isogenes which are present in both lineages A1xD1 and A1xD3 of V. longisporum could overcome these disadvantages. VlVTA2-1 has a similar size as V. dahliae VdVTA2 (1,701 nucleotides) and VlVTA2-2 resembles V. albo-atrum VaVTA2 (1,754–1,759 nucleotides). The difference in lengths between the two VTA2 isogenes is mainly due to differences in the third intron (Figs. 5a and 6a). We designed a specific primer pair (sVTA2-F/sVTA2-R) that amplifies only the third intron region (I3) of VTA2 gene. These primers bind to the conserved sequences in the third (E3) and fourth exon (E4) of VTA2 gene for all the three species (Figs. 6a and S5). With this primer pair, two bands of 315 and 380 bp, which are corresponding to VlVTA2-1 and VlVTA2-2, are amplified from genomic DNA of both V. longisporum lineages A1xD1 and A1xD3, whereas only a single band of 315 or 380 bp can be amplified from genomic DNA of V. dahliae or V. albo-atrum, respectively (Fig. 6b).
VTA2 is a barcode gene for specific recognition of V. longisporum hybrids. a Scheme for the structure of the third intronic region (I3) of VTA2 in three species. The specific primer pair (sVTA2-F/sVTA2-R) amplifying this intron is indicated by arrows. b PCR amplification with this primer pair results in two bands of 315 and 380 bp for both lineages A1xD1 and A1xD3 of the oilseed rape pathogen V. longisporum, whereas only a unique band of 315 or 380 bp is present in V. dahliae and V. albo-atrum, respectively
These results show that VTA2 can serve as barcode marker to discriminate between hybrid V. longisporum isolates and haploid V. dahliae or V. albo-atrum strains by a simple PCR reaction.
Combined diagnostic tools to identify virulent A1xD1 and avirulent A1xD3 V. longisporum hybrids among Verticillium species
The A1xD1 and A1xD3 V. longisporum hybrids from oilseed rape in Europe were originally identified by random amplified polymorphic DNA (RAPD)–PCR as groups lsp (A1xD1) or lsp* (A1xD3) which both produce longer conidia in comparison to the short spores of V. dahliae and V. albo-atrum. The lsp/A1xD1 group displays a high virulence on the oilseed rape Brassica napus by killing about 50 % plants after 42 days of infection, whereas the lsp*/A1xD3 group is significantly less virulent (Zeise and von Tiedemann 2002a, b). Like V. dahliae, both groups produce microsclerotia as resting structures. Depending on the medium, A1xD1 isolates can form elongated microsclerotia, whereas the A1xD3 isolates produce compact microsclerotia as V. dahliae (Fig. 7a). These morphological criteria vary under different environmental conditions and have to be supplemented by a set of molecular criteria (Fig. 7b). Two primer pairs specific to each isogene of the VTA2 barcode gene and to each rDNA type for V. longisporum discriminate between virulent A1xD1 (lsp) and avirulent A1xD3 (lsp*) after analyses in morphology of conidia and the resting structures. There are two PCR bands of 315 and 380 bp for both the virulent A1xD1 and the avirulent A1xD3, but only one PCR band of 315 or 380 bp for V. dahliae and V. albo-atrum, respectively (Fig. 6b). In a second step, the A1xD1 isolates can be discriminated from the A1xD3 isolates by the specific primer pair for the 18S rRNA gene of rDNA. A PCR band of 1.28 kb is unique for the A1xD1 isolates, whereas the genomes of the A1xD3 isolates represent a smaller PCR band of 0.44 kb that is also present in genomes of V. dahliae and V. albo-atrum (Fig. 2b). This combination between the VTA2 barcode gene analysis and the determination of the rDNA type might be a promising tool to distinguish haploid Verticillia from virulent A1xD1 (A1 rDNA) or avirulent A1xD3 (D3 rDNA) V. longisporum strains.
Criteria for discrimination of V. longisporum hybrids from each other and from V. dahliae and V. albo-atrum. a Morphological analysis for some representatives of V. longisporum, V. dahliae and V. albo-atrum. On the SXM medium mimicking xylem sap (Neumann and Dobinson 2003), two V. longisporum representative isolates produce long spores including the virulent isolate Vl43 (A1xD1) forms a mixture of elongate and compact microsclerotia whereas the avirulent one Vl32 (A1xD3) forms only compact microsclerotia comprising a group of many melanized round cells. V. dahliae produces short spores and compact microsclerotia with few melanized round cells, whereas V. albo-atrum also produces short spores but black hyphae as the resting structure. The same scale bars were indicated. b The summary of typical features as criteria for specific recognition of V. longisporum hybrids (A1xD1, A1xD3), V. dahliae and V. albo-atrum
Discussion
The two European V. longisporum hybrids A1xD1 and A1xD3 have both been isolated from oilseed rape but differ significantly in their pathogenicity towards this crop and can additionally be separated by their rDNA type. Both hybrid lineages have homogenized their rDNA to only one parental version. Two distinct rDNA types representing either the D3 V. dahliae parent or the A1 parent could be even isolated from the same site in Northern Germany. All A1xD1 V. longisporum isolates from Europe or California only represent the A1 rDNA type which is closer to V. albo-atrum than to V. dahliae. The D1 rDNA has not yet been found in any V. longisporum isolate. The loss of parental rDNA in V. longisporum hybrids might account for the described reduction from the diploid to the 1.8-fold ploidy of analyzed isolates. The recent classification of V. longisporum into the three different lineages A1xD1, A1xD2 and A1xD3 (Inderbitzin et al. 2011) includes the worldwide lineage A1xD1 with a broad host range including oilseed rape, cauliflower and Chinese cabbage. The A1xD2 hybrid is restricted to horseradish in Illinois, USA. A1xD3 is restricted to oilseed rape in Europe and Japan but infected plants display no disease symptoms (Zeise and von Tiedemann 2002a; Inderbitzin et al. 2011).
We analyzed here the rDNA types of A1xD1 and A1xD3 isolates and several gene pairs of putative regulators. All avirulent A1xD3 carry the D3 rDNA type; a virulent A1xD1 with a D1 rDNA was not found in the sample of 11 strains from California and Europe but all of them represented the A1 rDNA type. The analysis of several regulatory genes revealed that the alleles for strains D1 and D3 differ by several SNPs. V. dahliae strains with unusual hosts (like Vd14.01 growing on pistachio) could be more separated from other V. dahliae strains as the V. longisporum parent D1 (Fig. S2b) which has been grouped outside of V. dahliae (Inderbitzin et al. 2011).
Speciation by natural selection can favor alleles at different loci in a subpopulation in a specific environment distinct from the environment of another subpopulation. Similar selection might result in mutation-based speciation if subpopulations accumulate a different series of mutations, which is subsequently separated by ecological speciation (Schluter and Conte 2009). Fungal plant pathogens adapt rapidly to new hosts and are therefore interesting models for ecological speciation (Giraud et al. 2010). In nature, the hybrid V. longisporum infects only crops of the crucifer Brassicaceae family (Karapapa et al. 1997; Karapapa and Typas 2001; Zeise and von Tiedemann 2001; Barbara and Clewes 2003; Collins et al. 2003; Gladders et al. 2011), whereas V. dahliae and V. albo-atrum isolates infect more than 200 different plants except crucifers (Pegg and Brady 2002; Agrios 2005). The specialized inhabitation on crucifer hosts for V. longisporum might represent an ecologically dependent natural selection, where an interspecies hybrid is favored instead of its parents. At present, the hybrid can colonize ecological niches (crucifer crops) unavailable to the parents. It is currently unclear whether one parent had already accumulated some suitable specific mutations prior to hybridization, which supported the interaction to the new host or whether ecological speciation has only started after hybridization. In this study, we demonstrated that the V. dahliae VEL2 gene, an orthologue of A. nidulans VelB, has an increased accumulation of SNPs in isolates from linseed (Vd73) and pistachio (Vd14.01) in comparison to the other tested V. dahliae isolates. This increased SNP accumulation might play a role in host adaptation. The VelB protein interacts with VeA and acts as a light-dependent regulator of fungal development and secondary metabolism of A. nidulans (Bayram et al. 2008; Bayram and Braus 2012). VelB orthologues are conserved in different phytopathogenic fungi for virulence. Deletion of FfVel2 (VelB) in F. fujikuroi results in reduction in virulence on the host plant (Wiemann et al. 2010). Similarly, FgVelB deletion mutants of Fusarium graminearum are less virulent on wheat (Jiang et al. 2012). Vel2 (VelB) of pathogenic Verticillium species might also contribute to virulence on plant hosts. Accumulation of SNPs into VEL2 gene might be involved in fitness or enhanced virulence of the pathogens on specific plants.
The ecological separation of the parental habitat disconnects gene flow between a hybrid and both parents and the speciation of the hybrid becomes more feasible (Mallet 2007). The hybrid Zymoseptoria tritici was shown to be the result of a hybridization event of two divergent haploid individuals. The successful spread of the wheat pathogen Z. tritici is particularly accounted to the variation in gene density and recombination rate (Stukenbrock et al. 2012). The hybrid V. longisporum lineages separated from their parental strains by specialization on cruciferous hosts. It is unclear why the A1xD1 hybrids are significantly more virulent than the A1xD3 hybrids and still can be found on the host plant B. napus.
Polyploidy increases the number of duplicated sequences in the genome resulting in homologous recombination or gene conversion mechanisms that may lead to novel intergenic interactions (Wendel 2000). Organisms with a higher ploidy level have a higher potential for distinct beneficial alleles. The rate of adaptive evolution seems to increase with the ploidy level resulting in more beneficial mutations in polyploid populations (Otto and Whitton 2000). Interspecific hybridization might start with one genome from each parent (50:50), but recombination and gene conversion may eventually lead to unequal contributions (Mallet 2007). Interspecies hybrids can lead to dominant phenotypes resulting from the combination of the parental genomes. Hybridizations between closely-related fungal species play major roles in the generation of new species to invade new host plants (Brasier et al. 1999; Staats et al. 2005). Increasing the ploidy of V. longisporum by hybridization between V. dahliae and V. albo-atrum related species A1 might already change the spore size. This is supported by laboratory interspecies hybridization between V. dahliae and V. albo-atrum (Typas 1983). Some of these artificial diploid hybrids produced long spores (8.2 ± 0.2 μm) similar to the spores of V. longisporum. It is currently unclear whether the increased spore size represents a trait, which facilitates infection of cruciferous hosts.
Polyploidy results in duplicated genes where not necessarily both genes have to remain active. Both transcription factors encoding isogene pairs that we have analyzed are presumably expressed because we found for each a corresponding cDNA derived from the transcripts. Diploidization allows that one gene could retain its original function whereas the other copy may become epigenetically silenced or inactivated by mutations. Duplicated genes might also encounter recombination, which changes the genetic locus in a concerted evolution (Wendel 2000). Physical elimination of a parental genome-specific repeated DNA occurred in wheat in amphiploids in early generations. This may play a role in the initial stabilization of the nascent amphiploid plant (Han et al. 2005). Repeated genes might homogenize and the rDNA repeats are prominent examples where one variant can become dominant via mutations. Homogenization is caused by deletion events or continuous multiplication during unequal recombination. Single nucleotide polymorphisms are present in ribosomal genes with an unexpectedly high amount (Simon and Weiss 2008). Our analysis of V. longisporum showed that homogenization of rDNA genes can shift into both directions in nature further representing an early state of species formation.
Speciation is a highly dynamic process and might end in rehaploidization. Although most of V. longisporum isolates were reported to be genetically stable (Ingram 1968; Karapapa et al. 1997; Inderbitzin et al. 2011), two natural isolates of V. longisporum from rape and sugarbeet were forced to generate haploid segregants by treatment with the haploidizing agent p-fluorophenylalanine. These haploid segregants produced relatively short spores and their nuclear DNA content was halved resulting in similar values as V. dahliae or V. albo-atrum (Jackson and Heale 1985). Haploid Verticillium isolates were described from crucifers as recombinant isolates or secondary haploids, which were shown to be distinct from V. dahliae as well as from V. albo-atrum (Karapapa et al. 1997; Collins et al. 2003). It will be interesting to monitor the future development of V. longisporum to determine the kinetics of species formation, which might correlate with a reduction of the genome size. In addition, it is still an open question if factors have changed host specificity of the hybrid. This might include changes in gene dosage of specific genes, the accumulation of specific mutations in one copy of distinct isogene pairs or combinatory effects resulting from the combination of the two parental genomes. A further reduction of the genome size might disclose these host-specificity factors, which are still hidden in the 1.8 fold genome size of V. longisporum.
The interspecies hybrid V. longisporum includes two different lineages A1xD1 and A1xD3 in Europe which both grow on Brassicaceae but with a significantly different impact. A1xD1 hybrids are significantly more virulent than A1xD3 hybrids. We show here that virulent A1xD1 strains have lost the D1 parental rDNA, whereas avirulent A1xD3 strains have lost the A1 rDNA. This homogenization of rDNAs allows that both V. longisporum hybrids can be distinguished by their rDNA type in a single polymerase chain reaction. The amplification of 18S rRNA with the proposed primers results in a large band (1.28 kb) for virulent A1xD1 and a small band (0.44 kb) for avirulent A1xD3. The analysis of several regulatory gene pairs in V. longisporum resulted in a second tool for classification which allows the discrimination between the hybrids and haploid V. dahliae or V. albo-atrum strains. The PCR examination of the barcode marker VTA2 separates all hybrids by the resulting two bands from the single bands of V. dahliae and V. albo-atrum. Monitoring of the changes in the genomes of V. longisporum isolates will be an important part of a better understanding of host-specific growth of this fungus. Such diagnostic tools will support to oversee the dynamics of hybrid-mediated species formation of Verticillia as an emerging threat for agriculturally important crop plants.
References
Agrios GN (2005) Plant pathology. Elsevier, Burlington, USA
Barbara DJ, Clewes E (2003) Plant pathogenic Verticillium species: how many of them are there? Mol Plant Pathol 4:297–305
Bayram O, Braus GH (2012) Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24
Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506
Beser J, Hagblom P, Fernandez V (2007) Frequent in vitro recombination in internal transcribed spacers 1 and 2 during genotyping of Pneumocystis jirovecii. J Clin Microbiol 45:881–886
Brasier CM, Cooke DEL, Duncan JM (1999) Origin of a new Phytophthora pathogen through interspecific hybridization. Proc Natl Acad Sci USA 96:13589–13589
Calvo AM (2008) The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol 45:1053–1061
Clewes E, Edwards SG, Barbara DJ (2008) Direct molecular evidence supports long-spored microsclerotial isolates of Verticillium from crucifers being interspecific hybrids. Plant Pathol 57:1047–1057
Collado-Romero M, Jimenez-Diaz RM, Mercado-Blanco J (2010) DNA sequence analysis of conserved genes reveals hybridization events that increase genetic diversity in Verticillium dahliae. Fungal Biol 114:209–218
Collins A, Okoli CAN, Morton A, Parry D, Edwards SG, Barbara DJ (2003) Isolates of Verticillium dahliae pathogenic to crucifers are of at least three distinct molecular types. Phytopathology 93:364–376
de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, Zhang Z, Usami T, Lievens B, Subbarao KV, Thomma BP (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109:5110–5115
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T and Wilson A (2011) Geneious v5.4. Available from http://www.geneious.com/
Eastburn DM, Chang RJ (1994) Verticillium dahliae: a causal agent of root discoloration of horseradish in Illinois. Plant Dis 78:496–498
Fahleson J, Hu Q, Dixelius C (2004) Phylogenetic analysis of Verticillium species based on nuclear and mitochondrial sequences. Arch Microbiol 181:435–442
Ganley ARD, Kobayashi T (2007) Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res 17:184–191
Giraud T, Gladieux P, Gavrilets S (2010) Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol 25:387–395
Gladders P, Smith JA, Kirkpatrick L, Clewes E, Grant C, Barbara D, Barnes AV, Lane CR (2011) First record of Verticillium wilt (Verticillium longisporum) in winter oilseed rape in the UK. New Disease Reports 23:8
Glass NL, Kuldau GA (1992) Mating type and vegetative incompatibility in filamentous ascomycetes. Annu Rev Phytopathol 30:201–224
Goud JKC, Termorshuizen AJ, Gams W (2003) Morphology of Verticillium dahliae and V. tricorpus on semi-selective media used for the detection of V. dahliae in soil. Mycol Res 107:822–830
Han FP, Fedak G, Guo WL, Liu B (2005) Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGcIR-1a) in newly synthesized wheat allopolyploids. Genetics 170:1239–1245
Hastie AC (1973) Hybridization of Verticillium albo-atrum and Verticillium dahliae. Trans Br Mycol Soc 60:511–523
Heale JB, Karapapa VK (1999) The Verticillium threat to Canada’s major oilseed crop: canola. Can J Plant Pathol 21:1–7
Inderbitzin P, Davis RM, Bostock RM, Subbarao KV (2011) The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS One 6:e18260
Ingram R (1968) Verticillium dahliae Kleb. var. longisporum Stark: a stable diploid. Trans Br Mycol Soc 51:339–341
Isaac I (1967) Speciation in Verticillium. Annu Rev Phytopathol 5:201–222
Jackson CW, Heale JB (1985) Relationship between DNA content and spore volume in 16 isolates of Verticillium lecanii and 2 new diploids of Verticillium dahliae (=V. dahliae var. longisporum Stark). J Gen Microbiol 131:3229–3236
Jiang J, Yun Y, Liu Y, Ma Z (2012) FgVELB is associated with vegetative differentiation, secondary metabolism and virulence in Fusarium graminearum. Fungal Genet Biol 49:653–662
Karapapa VK, Bainbridge BW, Heale JB (1997) Morphological and molecular characterization of Verticillium longisporum comb. nov., pathogenic to oilseed rape. Mycol Res 101:1281–1294
Karapapa VK, Typas MA (2001) Molecular characterization of the host-adapted pathogen Verticillium longisporum on the basis of a group-I intron found in the nuclear SSU-rRNA gene. Curr Microbiol 42:217–224
Kim JT, Park IH, Lee HB, Hahm YI, YS H (2001) Identification of Verticillium dahliae and V. albo-atrum causing wilt of tomato in Korea. Plant Pathol J 17:222–226
Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV (2009) Diversity, pathogenicity and management of Verticillium species. Annu Rev Phytopathol 47:39–62
Klosterman SJ, Subbarao KV, Kang SC, Veronese P, Gold SE, Thomma BPHJ, Chen ZH, Henrissat B, Lee YH, Park J, Garcia-Pedrajas MD, Barbara DJ, Anchieta A, de Jonge R, Santhanam P, Maruthachalam K, Atallah Z, Amyotte SG, Paz Z, Inderbitzin P, Hayes RJ, Heiman DI, Young S, Zeng QD, Engels R, Galagan J, Cuomo CA, Dobinson KF, Ma LJ (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7:e1002137
Koike ST, Subbarao KV, Davis RM, Gordon TR, Hubbard JC (1994) Verticillium wilt of cauliflower in California. Plant Dis 78:1116–1121
Kolar M, Punt PJ, van den Hondel CAMJJ, Schwab H (1988) Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli LacZ fusion gene. Gene 62:127–134
Mallet J (2007) Hybrid speciation. Nature 446:279–283
Nagao H, Wakatabe D, Iijima T (1994) Difficulty to establish vegetative compatibility of Japanese isolates of Verticillium dahliae Kleb. using melanin-synthesis deficient mutants. J Gen Appl Microbiol 40:277–285
Neumann MJ, Dobinson KF (2003) Sequence tag analysis of gene expression during pathogenic growth and microsclerotia development in the vascular wilt pathogen Verticillium dahliae. Fungal Genet Biol 38:54–62
Odenbach D, Breth B, Thines E, Weber RWS, Anke H, Foster AJ (2007) The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol Microbiol 64:293–307
Okonechnikov K, Golosova O, Fursov M, Team U (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34:401–437
Pegg GF, Brady BL (2002) Verticillium wilts. CABI, Wallingford, Oxon, UK
Pramateftaki PV, Antoniou PP, Typas MA (2000) The complete DNA sequence of the nuclear ribosomal RNA gene complex of Verticillium dahliae: Intraspecific heterogeneity within the intergenic spacer region. Fungal Genet Biol 29:19–27
Qin QM, Vallad GE, Wu BM, Subbarao KV (2006) Phylogenetic analyses of phytopathogenic isolates of Verticillium spp. Phytopathology 96:582–592
Reinke J, Berthold G (1879) Die Zersetzung der Kartoffel durch Pilze. Untersuchungen aus dem Botanischen Laboratorium der Universität Göttingen 1:1–100
Rooney AP, Ward TJ (2005) Evolution of a large ribosomal RNA multigene family in filamentous fungi: Birth and death of a concerted evolution paradigm. Proc Natl Acad Sci USA 102:5084–5089
Saitou N, Nei M (1987) The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schluter D, Conte GL (2009) Genetics and ecological speciation. Proc Natl Acad Sci USA 106:9955–9962
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA 109:6241–6246
Schoustra SE, Debets AJM, Slakhorst M, Hoekstra RF (2007) Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet 3:e68
Simon UK, Weiss M (2008) Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol Biol Evol 25:2251–2254
Singh S, Braus-Stromeyer SA, Timpner C, Tran VT, Lohaus G, Reusche M, Knufer J, Teichmann T, von Tiedemann A, Braus GH (2010) Silencing of Vlaro2 for chorismate synthase revealed that the phytopathogen Verticillium longisporum induces the cross-pathway control in the xylem. Appl Microbiol Biotechnol 85:1961–1976
Staats M, van Baarlen P, van Kan JAL (2005) Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol Biol Evol 22:333–346
Stark C (1961) Das Auftreten der Verticillium-Tracheomykosen in Hamburger Gartenbaukulturen. Gartenbauwissenschaft 26:493–528
Steventon LA, Fahleson J, Hu Q, Dixelius C (2002) Identification of the causal agent of Verticillium wilt of winter oilseed rape in Sweden. V. longisporum Mycol Res 106:570–578
Studt L, Wiemann P, Kleigrewe K, Humpf HU, Tudzynski B (2012) Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl Environ Microbiol 78:4468–4480
Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH (2012) Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Natl Acad Sci USA 109:10954–10959
Subbarao KV, Chassot A, Gordon TR, Hubbard JC, Bonello P, Mullin R, Okamoto D, Davis RM, Koike ST (1995) Genetic-relationships and cross pathogenicities of Verticillium dahliae isolates from cauliflower and other crops. Phytopathology 85:1105–1112
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ and Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3
Typas MA (1983) Heterokaryon incompatibility and interspecific hybridization between Verticillium albo-atrum and Verticillium dahliae following protoplast fusion and microinjection. J Gen Microbiol 129:3043–3056
Vallad GE, Bhat RG, Koike ST, Ryder EJ, Subbarao KV (2005) Weedborne reservoirs and seed transmission of Verticillium dahliae in lettuce. Plant Dis 89:317–324
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, Humpf HU, Tudzynski B (2010) FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol 77:972–994
Yu JH, Butchko RAE, Fernandes M, Keller NP, Leonard TJ, Adams TH (1996) Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr Genetics 29:549–555
Zeise K, von Tiedemann A (2001) Morphological and physiological differentiation among vegetative compatibility groups of Verticillium dahliae in relation to V. longisporum. J Phytopathol 149:469–475
Zeise K, von Tiedemann A (2002a) Host specialization among vegetative compatibility groups of Verticillium dahliae in relation to Verticillium longisporum. J Phytopathol 150:112–119
Zeise K, von Tiedemann A (2002b) Application of RAPD-PCR for virulence type analysis within Verticillium dahliae and V. longisporum. J Phytopathol 150:557–563
Acknowledgments
The authors thank Andreas von Tiedemann (Göttingen) and Bart Thomma (Wageningen, Netherlands) for kindly providing most of the Verticillium isolates, Christiane Preiss for excellent technical assistance, and Irene Hampe for help in some experiments. We thank Harald Kusch for critically reading the manuscript. This work was funded by the Federal Ministry of Education and Research (BMBF) BioFung project and German Research Foundation (DFG) grant FOR546.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
PDF 15,000 kb
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Tran, V.T., Braus-Stromeyer, S.A., Timpner, C. et al. Molecular diagnosis to discriminate pathogen and apathogen species of the hybrid Verticillium longisporum on the oilseed crop Brassica napus . Appl Microbiol Biotechnol 97, 4467–4483 (2013). https://doi.org/10.1007/s00253-012-4530-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4530-1