Abstract
Biofuels synthesized from renewable resources are of increasing interest because of global energy and environmental problems. We have previously demonstrated production of higher alcohols from Escherichia coli using a 2-keto acid-based pathway. Here, we have compared the effect of various alcohol dehydrogenases (ADH) for the last step of the isobutanol production. E. coli has the yqhD gene which encodes a broad-range ADH. Isobutanol production significantly decreased with the deletion of yqhD, suggesting that the yqhD gene on the genome contributed to isobutanol production. The adh genes of two bacteria and one yeast were also compared in E. coli harboring the isobutanol synthesis pathway. Overexpression of yqhD or adhA in E. coli showed better production than ADH2, a result confirmed by activity measurements with isobutyraldehyde.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To meet the increasing global demand for energy and reduce the negative environmental impact, biofuels derived from renewable sources have become increasingly important. In particular, increasing attention has been paid to higher alcohols as potential substitutes for gasoline. Those alcohols possess a host of qualities making them more suitable as a liquid fuel than ethanol, including lower vapor pressure, lower hygroscopicity, and higher energy density. However, efficient production of these alcohols is challenging (Atsumi et al. 2008a; Cann and Liao 2008; Connor and Liao 2008; Inui et al. 2008; Shen and Liao 2008).
We previously devised a biosynthetic strategy to produce higher alcohols in Escherichia coli which takes advantage of the amino acid biosynthesis capability to produce various 2-keto acids and the broad substrate range of 2-keto acid decarboxylases (KDCs) and alcoholdehydrogenases (ADHs; Atsumi et al. 2008b). In this work, we investigate the effect of various ADHs which convert isobutyraldehyde to isobutanol in the isobutanol biosynthesis pathway. ADHs are oxidoreductases that catalyze the reversible oxidation of alcohols to aldehydes or ketones, with the corresponding reduction of nicotinamide adenine dinucleotide (NAD) or NAD phosphate (NADP; Jornvall et al. 1987). There is a versatile set of ADHs with distinct structural differences, different substrate specificities, and different use of cofactors and metals (Jornvall et al. 1987).
In a previous experiment (Atsumi et al. 2008b), isobutyraldehyde was detected only in trace amounts with the overexpression of ADH2 from Saccharomyces cerevisiae, indicating sufficient activity of ADH for reducing isobutyraldehyde to isobutanol in the strain (JCL260/pSA55/pSA69; Atsumi et al. 2008b). However, E. coli has six genes (adhE, adhP, eutG, yiaY, yqhD, and yjgB) coding for various ADHs. It was unclear how much these ADHs contributed to the isobutanol production. In particular, YqhD has a preference for alcohols longer than C3, and it does not show detectable enzymatic activity when tested with short-chain alcohols as substrates (Sulzenbacher et al. 2004). However, its activity for isobutyraldehyde has not been reported. In this work, we first investigated whether ADHs of E. coli play a role in the isobutanol production (Perez et al. 2008). To improve the isobutanol production further, we also compared the activities of Adh2 from S. cerevisiae and AdhA from Lactococcus lactis for isobutanol production in E. coli. These ADHs along with YqhD from E. coli were purified and characterized in vitro and in vivo.
Materials and methods
Reagents
Restriction enzymes and Antarctic phosphatase were from New England Biolabs (Ipswich, MA, USA). Rapid DNA ligation kit was from Roche (Mannheim, Germany). KOD DNA polymerase was from EMD Chemicals (San Diego, CA, USA). Oligonucleotides were from Operon (Huntsville, AL, USA).
Strains and plasmids
A list of the strains and plasmids used is given in Table 1. All oligonucleotides are listed in Table 2. The yqhD gene was deleted by P1 transduction from a strain JW2978 (Baba et al. 2006). The yqhD deletion was confirmed with polymerase chain reaction (PCR). To clone kivd which encodes KDC, we used genomic DNA of L. lactis (ATCC) as PCR template with a pair of primers A96 and A97. PCR products were digested with Acc65I and SphI and cloned into pZE12-luc (Lutz and Bujard 1997) cut with the same enzyme, creating pSA129. To clone yqhD, genomic DNA of E. coli MG1655 was used as PCR template with a pair of primers K453 and A257. PCR products were digested with SphI and XbaI and cloned into pSA129 cut with the same enzyme, creating pSA138. To clone adhA, genomic DNA of L. lactis was used as PCR template with a pair of primers A121 and A122. PCR products were digested with SphI and XbaI and cloned into pSA55 cut with the same enzyme, creating pSA65.
For protein overexpression and purification, ADH2, adhA, and yqhD were amplified with primers A287 and A288, A289 and A290, and A292 and A299, respectively. PCR products were digested with BamHI and SalI and cloned into pETDuet-1 (Novagen (Madison, WI, USA)) cut with the same enzymes, creating pTW2, pTW3, and pTW4.
Medium and culture conditions for isobutanol production
M9 medium containing 36 g/L glucose, 5 g/L yeast extract, 100 μg/ml ampicillin, 30 μg/ml kanamycin, and 1,000th dilution of Trace Metal Mix A5 (2.86 g H3BO3, 1.81 g MnCl2⋅4H2O, 0.222 g ZnSO4⋅7H2O, 0.39 g Na2MoO4⋅2H2O, 0.079 g CuSO4⋅5H2O, 49.4 mg Co(NO3)2⋅6H2O per liter water) was used for cell growth. Preculture in test tubes containing 3 ml of medium was performed at 37°C overnight on a rotary shaker (250 rpm). Overnight culture was diluted 1:100 into 20 ml of fresh medium in a 250-ml screw cap conical flask. Cells were grown at 37°C for 3 h, followed by adding 0.1 mM isopropyl-β-d-thio-galactoside (IPTG). Production was performed at 30°C on a rotary shaker (250 rpm) for 24 h. Gas chromatography-flame ionization detector analysis is carried out as previously described (Atsumi et al. 2008b).
Protein purification
YqhD was overexpressed from pTW2 in E. coli BL21 Star™ (DE3) (Invitrogen (Carlsbad, CA, USA)). Adh2 and AdhA were overexpressed from pTW3 and pTW4, respectively, in BL21-CodonPlus(DE3)-RIL-X (Stratagene, Cedar Creek, TX, USA). Overexpressed proteins were purified with Ni-NTA Spin Columns (Qiagen (Valencia, CA, USA)). Protein concentrations were determined by the Bradford assay (Bio-Rad (Hercules, CA, USA)).
Protein solubility assay
The strains with pTW2, pTW3, and pTW4 were grown to OD600 value of 0.4–0.6 in 5 mL Luria–Bertani medium at 37°C, followed by adding 1 mM IPTG. Protein overexpression was performed at 30°C for 4 h. The cells were centrifuged, resuspended in 250 μl BugBuster Protein Extraction Reagent (Novagen, San Diego, CA, USA), and incubated at room temperature for 20 min for cell lysis. To separate soluble and insoluble proteins, the samples were centrifuged for 50 min (13,000 rpm, 4°C). Supernatant was taken as a soluble sample. The cell pellets were resuspended by 250 μl 8 M urea and centrifuged. Supernatant was taken as an insoluble sample. Soluble and insoluble protein samples were visually assessed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme activity assay
The reaction mixture contained 50 mM 3-(N-morpholino)propanesulfonic acid buffer, pH 7.0, 0.25 mM NAD(P)H, a defined aldehyde (acetaldehyde or isobutyraldehyde) as substrate, and purified ADH (100 nM). The mixture was incubated at 37°C, and NAD(P)H oxidation was determined at 340 nm using a spectrophotometer (Beckman Coulter DU800). One unit of enzyme activity was defined as the amount of protein that oxidizes 1 pmol of NAD(P)H/min at 37°C. The K m values for different aldehydes and the V max were extrapolated after nonlinear regression of the experimental points with Gauss–Newton method using Matlab.
Results
Effects of E. coli ADHs on isobutanol production
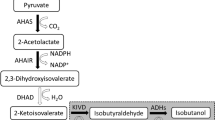
In a previous study to produce isobutanol, the carbon source was converted to 2-ketoisovalerate, through overexpressed AlsS (Bacillus subtilis), IlvC (E. coli), and IlvD (E. coli; Atsumi et al. 2008b; Fig. 1). The resulting 2-ketoisovalerate is then converted to isobutanol using KDC (de la Plaza et al. 2004) and ADH (Fig. 1). In addition, six genes that are involved in byproduct formation were deleted (Atsumi et al. 2008b). The isobutanol strain (JCL260/pSA55/pSA69) produced more than 20 g/L in 112 h (Atsumi et al. 2008b). The production host, JCL260, has ∆adhE to reduce ethanol production. Thus, the strain still has five genes (adhP, eutG, yiaY, yqhD, and yjgB) on the genome coding for ADHs.
To evaluate the effects of these ADHs for isobutanol production, isobutanol production was tested without overexpression of ADH from plasmids (Fig. 2a). This strain (JCL260/pSA69/pSA129) produced 7.4 g/L isobutanol after 24 h which is similar to those of the strain expressing ADH2 of S. cerevisiae from pSA55 (Fig. 2a and Table 1). Since there is no adh gene on plasmids, the E. coli ADHs are responsible for the conversion of isobutyraldehyde to isobutanol.
Isobutanol production with YqhD and Adh2. The cells were grown in M9 medium containing 5 g/L yeast extract and 36 g/L of glucose in shake flasks at 30°C with 0.1 mM IPTG for 24 h. Optical density at 600 nm (OD600) of all strains described here at 24 h was ∼6.0. Overexpressed and deleted genes are indicated below the axis. Plasmid ADH2 (triangle) and (plus sign) indicate pSA129 and pSA55, respectively. a Isobutanol production. b Isobutyraldehyde production
Elimination of yqhD decreases isobutanol production
Among these ADHs, YqhD is known to have a preference for longer-chain substrates. YqhD is an NADPH/Zn2+-dependent enzyme (Sulzenbacher et al. 2004). To identify the effect of YqhD for isobutyraldehyde reductase activity, the yqhD gene was deleted from the genome. Isobutanol production with ∆yqhD decreased by 80% (Fig. 2a). Isobutyraldehyde was accumulated (Fig. 2b). These results indicate that YqhD contributed significantly to the isobutyraldehyde reductase activity in E. coli.
Overexpression of ADH2, adhA, and yqhD
To study the effects of overexpression of various ADHs, we cloned three adh genes on plasmids. First, overexpression of ADH2 from S. cerevisiae did not increase isobutanol production compared to the strain without overexpression (Fig. 2a). A deletion of the yqhD gene from this strain resulted in a decrease of isobutanol production to 3.8 g/L from 6.8 g/L and the accumulation of isobutyraldehyde (2.7 g/L; Fig. 2b). However, this was higher than the production (1.4 g/L) from the strain (∆yqhD) without overexpression of ADH2 (Fig. 2a). This result suggests that Adh2 has some activities toward isobutyraldehyde but is not as efficient as YqhD in E. coli.
The adhA gene of L. lactis was identified by genomic sequencing (Bolotin et al. 2001). AdhA of L. lactis has not been characterized biochemically. However, for isobutanol production, we used KDC encoded by the kivd gene from L. lactis which shows a broad-range substrate specificity (Atsumi et al. 2008b; de la Plaza et al. 2004); therefore, we expected that AdhA could have a broad-range substrate specificity. Overexpression of adhA from pSA65 (Table 1) increased isobutanol production slightly in the yqhD + strain (Fig. 3a). Interestingly, a deletion of the chromosomal yqhD gene in the presence of the overexpressed AdhA did not reduce isobutanol production (Fig. 3a). In addition, isobutyraldehyde was detected only in trace amounts (data not shown), indicating that the activity of AdhA toward isobutyraldehyde is relatively high.
Isobutanol production with overexpressed YqhD and AdhA. Comparison of isobutanol production by each combination of overexpression and deletion of Adhs. Overexpressed and deleted genes are indicated below the axis. OD600 of all strains described here at 24 h was ∼6.0. a The effects of AdhA on isobutanol production. Plasmid adhA (triangle) and (plus sign) indicate pSA129 and pSA65, respectively. b The effects of YqhD on isobutanol production. Plasmid yqhD (triangle) and (plus sign) indicate pSA129 and pSA138, respectively
Finally, we tested whether overexpression of yqhD could improve the isobutanol production. In our system, the yqhD gene was cloned into a high copy plasmid (∼70 copies; pSA138, Table 1). Isobutanol production was measured with or without overexpression of yqhD. Unexpectedly, both strains showed the similar productivity (Fig. 3b). To eliminate the possibility for no expression of yqhD from the plasmids, we tested isobutanol production using the strain with ∆yqhD on the genome. The deletion did not change isobutanol production (Fig. 3b), suggesting that YqhD was expressed well from plasmids. The results indicate that chromosomal yqhD expression has saturated the isobutanol production capability of the strain.
Characterization of ADHs
We further assessed the efficiency of expression and the solubility of three ADHs in E. coli. As shown in Fig. 4, Adh2 is highly prone to misfolding and aggregation in the heterologous host E. coli. While significant amounts of soluble protein of YqhD and AdhA were detected, nearly all of the overexpressed Adh2 was found in the insoluble fractions of the cell lysate (Fig. 4).
To purify these proteins, Adh2, AdhA, and YqhD were expressed from a His tag plasmid (pTW2, pTW3, and pTW4) and purified as described in “Materials and methods.” The kinetic parameters were measured for these proteins by monitoring NAD(P)H oxidation. The K m values for acetaldehyde and isobutyraldehyde and the k cat are reported in Table 3. YqhD showed the highest K acetaldehydem value compared to AdhA and Adh2 (Table 3). YqhD showed lower K m value for isobutyraldehyde than that for acetaldehyde and the similar k cat for both substrates (Table 3). This is consistent with the data of previous report (Sulzenbacher et al. 2004). The K acetaldehydem and K isobutyraldehydem values of Adh2 are 0.14 and 385 mM, respectively, indicating that Adh2 prefers a short-chain substrate (Table 3). This result is consistent with isobutanol production described above. As predicted by sequence analysis, AdhA from L. lactis did not show any detectable enzymatic activity when tested with NADPH as a cofactor (Table 3). AdhA showed relatively small substrate preference towards acetaldehyde, but it exhibited the highest activity for isobutyraldehyde among the three enzymes tested. The glycolysis pathway produces two NADH per glucose but does not produce NADPH. Therefore, NADH-dependent enzymes should be preferred for isobutanol production. These results suggest that AdhA is most suitable for isobutanol production in an engineered pathway, consistent with the data shown in Fig. 3.
Discussion
The last step of isobutanol production in E. coli was examined by employing various ADHs. We found that YqhD contributed significantly to the isobutyraldehyde reductase activity in E. coli. We further compared three enzymes for the last step and found that AdhA from L. lactis showed the highest isobutyraldehyde reductase activity. The in vitro enzyme assay showed that AdhA was NADH-dependent, consistent with the prediction based on sequence. AdhA contains a highly conserved sequence Gly–X–Gly–X–X–Gly (where X is any amino acid) in many NADH-binding domains (Scrutton et al. 1990). Adh2 from S. cerevisiae was highly prone to misfolding and aggregation in the heterologous host E. coli.
There is strong incentive to devise a synthetic pathway for the production of valuable metabolites in a user-friendly organism. In this regard, E. coli is a well-characterized microorganism with a set of readily available tools for genetic manipulation, and its physiological regulation is well-studied. However, the expression of heterologous proteins may result in inefficient translation and misfolding (Baneyx and Mujacic 2004). To achieve high productivity of the target products, it is desirable to seek enzymes that are compatible to the host. Nearly all of the overexpressed Adh2 was found in the insoluble fraction of the cell lysate, indicating that Adh2 is highly prone to misfolding and aggregation when expressed in E. coli. Although a number of strategies have been developed to improve the folding of a protein (Fisher et al. 2006; Fowler et al. 2005), the comparison of several enzymes from a broad range of possibilities allowed us to identify an ideal enzyme for our purpose.
We found YqhD and AdhA were active toward isobutyraldehyde, but Adh2 shows a high K isobutyraldehydem value. For YqhD, it is consistent with the previous report that YqhD has a preference for longer-chain substrates (Perez et al. 2008; Sulzenbacher et al. 2004). The crystal structure indicates that YqhD has an elongated shape of the active site (Sulzenbacher et al. 2004). The large size of the active site is in agreement with the enzyme's preference for long substrates. For AdhA, in the absence of actual crystallographic data, we cannot discuss the specific mechanisms responsible for the broad-range substrate binding, but it might have a large active site cavity. Possibilities still exist that alternative ADHs from other organisms might show better activity toward isobutyraldehyde in E. coli. The crystal structures of more than 100 ADHs have been elucidated (e.g., Schwarzenbacher et al. 2004; Valencia et al. 2004). It is possible to use the structural information to identify ADHs that have large active sites. The use of the structural data could facilitate such exploration to identify better enzymes.
References
Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC (2008a) Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng 10(6):305–311
Atsumi S, Hanai T, Liao JC (2008b) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451(7174):86–89
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:E1–E11
Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22(11):1399–1408
Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11(5):731–753
Cann AF, Liao JC (2008) Production of 2-methyl-1-butanol in engineered Escherichia coli. Appl Microbiol Biotechnol 81(1):89–98
Connor MR, Liao JC (2008) Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl Environ Microbiol 74(18):5769–5775
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12):6640–6645
de la Plaza M, Fernandez de Palencia P, Pelaez C, Requena T (2004) Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett 238(2):367–374
Fisher AC, Kim W, DeLisa MP (2006) Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci 15(3):449–458
Fowler SB, Poon S, Muff R, Chiti F, Dobson CM, Zurdo J (2005) Rational design of aggregation-resistant bioactive peptides: reengineering human calcitonin. Proc Natl Acad Sci U S A 102(29):10105–10110
Inui M, Suda M, Kimura S, Yasuda K, Suzuki H, Toda H, Yamamoto S, Okino S, Suzuki N, Yukawa H (2008) Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol 77(6):1305–1316
Jornvall H, Persson B, Jeffery J (1987) Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur J Biochem 167(2):195–201
Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25(6):1203–1210
Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC (2008) Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem 283(12):7346–7353
Schwarzenbacher R, von Delft F, Canaves JM, Brinen LS, Dai X, Deacon AM, Elsliger MA, Eshaghi S, Floyd R, Godzik A et al (2004) Crystal structure of an iron-containing 1,3-propanediol dehydrogenase (TM0920) from Thermotoga maritima at 1.3 A resolution. Proteins 54(1):174–177
Scrutton NS, Berry A, Perham RN (1990) Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343(6253):38–43
Shen CR, Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng 10(6):312–320
Sulzenbacher G, Alvarez K, Van Den Heuvel RH, Versluis C, Spinelli S, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M (2004) Crystal structure of E. coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J Mol Biol 342(2):489–502
Valencia E, Larroy C, Ochoa WF, Pares X, Fita I, Biosca JA (2004) Apo and Holo structures of an NADPH-dependent cinnamyl alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol 341(4):1049–1062
Acknowledgments
This work was partially supported by the UCLA-DOE Institute for Genomics and Proteomics. T. W. is supported by a Graduate Students Study Abroad Program from a National Science Council of R.O.C (Taiwan) (NSC-096-2917-I-007-102).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Atsumi, S., Wu, TY., Eckl, EM. et al. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol 85, 651–657 (2010). https://doi.org/10.1007/s00253-009-2085-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2085-6