Abstract
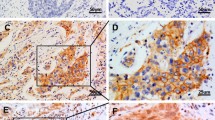
The aim of this study was to detect FOXC1 expression in human non-small cell lung cancer (NSCLC) and to analyze its association with prognosis of NSCLC patients. Expressional levels of FOXC1 mRNA and protein in 30 cases of NSCLC and corresponding non-tumor tissue samples were examined by quantitative real-time PCR and Western blotting. Immunohistochemistry was performed to detect the expression of FOXC1 in 125 NSCLC tissues. We found that the expression levels of FOXC1 mRNA and protein in NSCLC tissues were significantly higher than those in corresponding non-tumor tissues. High-level FOXC1 expression was correlated with poor tumor differentiation, tumor–node–metastasis stage, and lymph node metastasis. Patients with high expression levels of FOXC1 showed lower overall survival rate than those with low expression levels. Multivariate analysis showed that high FOXC1 protein expression was an independent prognostic factor for NSCLC patients. Our study suggests that over-expression of FOXC1 may play an important role in the progression of NSCLC, and FOXC1 expression may offer a valuable marker for predicting the outcome of patients with NSCLC.




Similar content being viewed by others
References
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi:10.3322/CA.2007.0010.
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–50. doi:10.1200/JCO.2005.05.2308.
Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–59. doi:10.1038/nrc2223.
Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL. The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev Biol. 1999;211:306–22.
Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–96.
Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerbäck S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld–Rieger anomaly. Am J Hum Genet. 1998;63:1316–28.
Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z, Zhu S, Jiang H, Shao Z, Huang B, Lu J. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res Treat. 2012;131:65–73.
Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, Barroilhet LM, Ng AS, Wong RR, Wang VW, Mok SC, Smith DI, Berkowitz RS, Wong YF. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–45. doi:10.1002/ijc.26060.
Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP, Ye X, Liu B, Elashoff D, Hoon DS, Walter MA, Martens JW, Richardson AL, Giuliano AE, Cui X. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70:3870–6. doi:10.1158/0008-5472.
Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V, Antosiewicz JE, Argani P, Halushka MK, Thomson JA, Pharoah P, Porgador A, Sukumar S, Parsons R, Richardson AL, Stampfer MR, Gelman RS, Nikolskaya T, Nikolsky Y, Polyak K. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105:14076–81. doi:10.1073/pnas.0805206105.
Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP, Lee AV, Lin X, Bagaria SP, Giuliano AE, Cui X. FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-κB signaling. Oncogene. 2012;31:4798–802. doi:10.1038/onc.2011.635.
Ray PS, Bagaria SP, Wang J, Shamonki JM, Ye X, Sim MS, Steen S, Qu Y, Cui X, Giuliano AE. Basal-like breast cancer defined by FOXC1 expression offers superior prognostic value: a retrospective immunohistochemical study. Ann Surg Oncol. 2011;18:3839–47.
Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, Hollier BG, Ram PT, Lander ES, Rosen JM, Weinberg RA, Mani SA. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi:10.1073/pnas.1004900107.
Sobin LH, Fleming ID. TNM classification of malignant tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–4.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Piperi C, Vlastos F, Farmaki E, Martinet N, Papavassiliou AG. Epigenetic effects of lung cancer predisposing factors impact on clinical diagnosis and prognosis. J Cell Mol Med. 2008;12:1495–501. doi:10.1111/j.1582-4934.2008.00309.x.
Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–28. doi:10.2353/ajpath.2009.080874.
Yang S. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–33. doi:10.1038/nrg2218.
Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi:10.1002/ijc.23605.
Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2012. doi:10.1002/hep.26029.
Peraldo-Neia C, Migliardi G, Mello-Grand M, Montemurro F, Segir R, Pignochino Y, Cavalloni G, Torchio B, Mosso L, Chiorino G, Aglietta M. Epidermal growth factor receptor (EGFR) mutation analysis, gene expression profiling and EGFR protein expression in primary prostate cancer. BMC Cancer. 2011;11:31. doi:10.1186/1471-2407-11-31.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wei, LX., Zhou, RS., Xu, HF. et al. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumor Biol. 34, 941–946 (2013). https://doi.org/10.1007/s13277-012-0629-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0629-3