Abstract
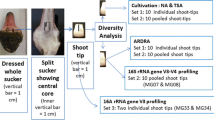
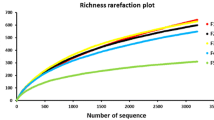
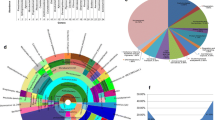
Diversity of bacterial endophytes associated with grapevine leaf tissues was analyzed by cultivation and cultivation-independent methods. In order to identify bacterial endophytes directly from metagenome, a protocol for bacteria enrichment and DNA extraction was optimized. Sequence analysis of 16S rRNA gene libraries underscored five diverse Operational Taxonomic Units (OTUs), showing best sequence matches with γ-Proteobacteria, family Enterobacteriaceae, with a dominance of the genus Pantoea. Bacteria isolation through cultivation revealed the presence of six OTUs, showing best sequence matches with Actinobacteria, genus Curtobacterium, and with Firmicutes genera Bacillus and Enterococcus. Length Heterogeneity-PCR (LH-PCR) electrophoretic peaks from single bacterial clones were used to setup a database representing the bacterial endophytes identified in association with grapevine tissues. Analysis of healthy and phytoplasma-infected grapevine plants showed that LH-PCR could be a useful complementary tool for examining the diversity of bacterial endophytes especially for diversity survey on a large number of samples.
Similar content being viewed by others
References
Araujo, W.L., J. Marcon, W. Maccheroni, J.D. van Elsas, Jr., J.W.L. van Vuurde, and J.L. Azevedo. 2002. Diversità of endophytic bacterial populations and their interaction with Xylella fastidiosa citrus plants. Appl. Environ. Microbiol. 68, 4906–4914.
Bell, C.R., G.A. Dickie, W.L.G. Harvey, and J.W.Y.F. Chan. 1994. Endophytic bacteria in grapevine. Can. J. Microbiol. 41, 46–53.
Ben-Dov, E., O.H. Shapiro, N. Siboni, and A. Kushmaro. 2006. Advantage of using inosine at the 3′ terminal of 16S rRNA gene universal primers for the study of microbial diversity. Appl. Environ. Microbiol. 72, 6902–6909.
Berg, G., A. Krechel, M. Ditz, R. Sikora, A. Ulrich, and J. Hallman. 2005. Endophytic and ectophytic potato-associted bacterial commmunities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 51, 215–229.
Bonaterra, A., M. Mari, L. Casalini, and E. Montesinos. 2003. Biological control of Monilinia laxa and Rizophus stolonifer in postharvest of stone fruit by Pantoea agglomerans EPS125 and putative mechanisms of antagonism. Int. J. Food Microbiol. 84, 93–104.
Brooks, D.S., C.F. Gonzalez, D.N. Appel, and T.H. Filer. 1994. Evaluation of endophytic bacteria as potential biological control agents for oak wilt. Biol. Control. 4, 373–381.
Brusetti, L., S. Borin, D. Mora, A. Rizzi, N. Raddadi, C. Sorlini, and D. Daffonchio. 2006. Usefulness of length heterogeneity-PCR for monitoring lactic acid bacteria succession during maize ensiling. FEMS Microbiol. Ecol. 56, 154–164.
Cankar, K., H. Kraigher, M. Ravinkar, and K.M. Rupnik. 2005. Bacterial endophytes from seeds of Norway spruce (Picea albis L. Karst). FEMS Microbiol. Lett. 244, 341–345.
Chelius, M.K. and E.W. Triplett. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41, 252–263.
Conn, V. and C.M.M. Franco. 2004. Analysis of the endophytic actinobacterial population in roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl. Environ. Microbiol. 70, 1787–1794.
Curtis, T.P., W.T. Sloan, and J.W. Scannel. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99, 10494–10499.
Davis, R.E. and E.L. Dally. 2001. Revised subgroup classification of group 16SrV phytoplasmas and placement of Flavescence dorèe-associated phytoplasmas in two distinct subgroups. Plant Dis. 85, 790–797.
Deng, S. and C. Hiruki. 1991. Genetic relatedness between two non-culturable myciplasmalike organisms revealed by nucleic acid hybridyzation and polymerase chain reaction. Phytopathology 81, 1475–1479.
Dent, K.C., J.R. Stephen, and W.E. Finch-Savage. 2004. Molecular profiling of microbial community associated with seeds of Beta vulgaris subsp. vulgaris (Sugar beet). J. Microbiol. Methods 56, 17–26.
Dunbar, J., S. Takala, S.M. Barns, A.J. Davis, and C.R. Kuske. 1999. Levels of bacterial community diversity in four arid soil compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65, 1662–1669.
Faoro, F. 2005. Why do grapevine phytoplasmas escape electron microscopists? Petria 15(1/2), 99–101.
Ferreira, A., M.C. Quecine, P.T. Lacava, S. Oda, J.L. Azevedo, and W.L. Araujo. 2008. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 287, 8–14.
Frank, J.A., C.I. Reich, S. Sharma, J.S. Weisbaum, B.A. Wilson, and G.J. Olsen. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470.
Gray, E.J. and D.L. Smith. 2005. Intracellular and extracellular PGPR: commonalities and distinction in the plant bacterium signalling processes. Soil Biol. Biochem. 37, 395–412.
Gutierrez-Zamora, M.L. and E. Martínez-Romero. 2001. Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J. Biotechnol. 91, 117–126.
Haiwen, L., J. Luo, J.K. Hemphill, J.T. Wang, and J.H. Gould. 2001. A rapid and high yielding DNA Miniprep for cotton (Gossypium spp.). Plant Mol. Biol. Rep. 19, 183a–183e.
Hallmann, J., A. Quadt-Hallmann, W.F. Mahaffee, and J.W. Kloepper. 1997. Bacterial endophytes in agicolture crops. Can. J. Microbiol. 43, 895–914.
Hengstmann, U., K.J. Chin, P.H. Janssen, and W. Liesack. 1999. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numericcaly abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65, 5050–5058.
Hurek, T., B. Reinhold-Hurek, M. Van Montagu, and E. Kellemberg. 1994. Root colonization and systemic spreeding of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 176, 1913–1923.
Huws, S.A., J.E. Edwards, E.J. Kim, and N.D. Scollan. 2007. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems, J. Microbiol. Methods 3, 565–569.
Idris, R., R. Trifonova, M. Puschenreitr, W.W. Wenzel, and A. Sessitch. 2004. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 70, 2667–2677.
Ishimaru, C.A.E., E. Klobs, and R.R. Brubaker. 1988. Multiple antibiotic production by Erwinia herbicola. Phytopatology 78, 746–750.
Jacobs, M.J., W.M. Bugbee, and D.A. Gabrielson. 1985. Enumeration, location and characterization of endophytic bacteria within sugar beet roots. Can. J. Bot. 63, 1262–1265.
Jensen, M.A., J.A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59, 945–952.
Jiao, J.Y., H.X. Wang, Y. Zeng, and Y.M. Shen. 2006. Enrichment for microbes living in association with plant tissues. J. Appl. Microbiol. 100, 830–837.
Kado, C.I. 1992. Plant pathogenic bacteria, p. 660–662. In A. Ballows, G.G. Truper, M. Dworkin, W. Harden, and K.H. Schleifer (eds.), The prokaryotes, Springerverlang, New York, N.Y., USA.
Kaiser, O., A. Pühler, and W. Selbitschka. 2001. Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb. Ecol. 42, 136–149.
Kaul, S., M. Wani, K.L. Dhar, and M.K. Dhar. 2008. Production and GC-MS trace analysis of methyl eugenol from endophytic isolate of Alternaria from rose. Ann. Microbiol. 58, 443–446.
Kumar S., J. Dudley, M. Nei, and K. Tamura. 2008. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9, 299–306.
Lane, D.J. 1991. 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics, p. 133. Modern microbiological methods. In E. Stackebrandt and M. Goodfellow (eds.), J. Wiley & Sons, Chichester, UK.
Lee, I.-M., D.E. Gundersen-Rindal, R. Davis, and I.M. Bartoszyk. 1998. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Bacteriol. 48, 1153–1169.
Marzorati, M., A. Alma, L. Sacchi, M. Pajoro, S. Palermo, L. Brusetti, N. Raddadi, A. Balloi, R. Tedeschi, E. Clementi, S. Corona, F. Quaglino, P.A. Bianco, T. Beninati, C. Bandi, and D. Daffonchio. 2006. A novel Bacteroidetes symbiont is localized in Scaphoideus titanus, the insect vector of Flavescence Dorée in Vitis vinifera. Appl. Environ. Microbiol. 72, 1467–1475.
Mills, D.K., J.A. Entry, P.M. Gillevet, and K. Mathee. 2007. Assessing microbial community diversity using amplicon length heterogeneity polymerase chain reaction. Soil Sci. Soc. Am. J. 71, 572–578.
Muyzer, G., S. Hottentrager, A. Teske, and C. Wawer. 1996. Denaturing gradient gel eletrophoresisfor PCR amplified 16s rRNA gene a new molecular approach to analyze the genetic diversity of mixed mcrobial communities, p. 1–23. In A.D.L. Akkermans, D.J. van Elsas, and F.J. Bruijin (eds.), Molecular Microbial Ecology Manual 3.4.4. Kluwer academic publisher, Dordrecht, The Netherlands.
Ortmann, I., U. Conrath, and B.M. Moerschbacher. 2006. Exopolysaccharides of Pantoea agglomerans have different priming and eliciting activities in suspension-cultured cells of monocots and dicots. FEMS Lett. 580, 4491–4494.
Prince, J.P., R.E. Davis, T.K. Wolf, I.M Lee, B.D. Mogen, E.L. Dally, A. Bartaccini, R. Credi, and M. Barba. 1993. Molecular detection of diverse mycoplasma like organisms (MLOs) associated with grapevine yellows and their classification with aster yellows MLOs. Phytopathology 83, 1130–1137.
Pusey, P.L., V.O. Stockwell, and D.R. Rudell. 2008. Antibiosis and acidification by Pantoea agglomerans strain E325 may contribute to suppression of Erwinia amylovora. Phytopathology 98, 1136–1143.
Raupach, G.S. and J.W. Kloepper. 1998. Mixtures of plant-growth promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology 88, 1158–1164.
Raupach, G.S. and J.W. Kloepper. 2000. Biocontrol of cucumber diseases in the field by plant-growth promoting rhizobacteria with and without methyl bromide fumigation. Plant Dis. 84, 1073–1075.
Ritchie, N.J., M.E. Schutter, R.P. Dick, and D.D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66, 1668–1675.
Schneider, B., E. Seemuller, C.D. Smart, and B.C. Kirkpatrick. 1995. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas, p. 369–380. In S. Razin and J.G. Tully (eds.), Molecular microbial and diagnostic procedures in mycoplasmology. Academic press, San Diego, USA.
Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2001. Analysis of endophytic bacteria in three potato cultivars. Abstr. 9th Meet. Int. Soc. Mol. Ecol., Abstr TU.052.
Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2002. Cultivation-independent population of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39, 23–32.
Shuman, S. 1994. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J. Biol. Chem. 269, 32678–32684.
Sun, L., F. Qiu, X. Zhang, X. Dai, X. Dong, and W. Song. 2008. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequences analysis. Microb. Ecol. 55, 415–424.
Taberlet, P., L. Gielly, G. Pautou, and J. Bouvet. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109.
Taechowisan, T. and S. Lumyong. 2003. Activity of endophytic actimomycetes from roots of Zingiber officinale and Alpinia galanga against phytopathogenic fungi. Ann. Microbiol. 53, 291–298.
Ulrich, K., A. Ulrich, and D. Ewald. 2008. Diversity of endophytic bacterial communities in poplar grown under field condition. FEMS Microbiol. Ecol. 63, 169–180.
Vega, F.E., M. Pava-Ripoll, F. Posada, and Y.S. Buyer. 2005. Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 45, 371–380.
Vidaver, A.K. 1982. The plant pathogenic corynebacteria. Annu. Rev. Microbiol. 36, 495–517.
Vorwerk, S., D. Martinez-Torres, and A. Forneck. 2007. Pantoea agglomerans-associated bacteria in grape phylloxera (Daktulosphaira vitifoliae, Fitch). Agr. Forest. Entomol. 9, 57–64.
Whitesides, S.K. and R.A. Spotts. 1991. Frequency, distribution, and characteristics of endophytic Pseudomonas syringe in pear trees. Phytopathology 81, 453–457.
Wilson, D. 1995. Endophyte: the evolution of a term, and clarification of its use and definition. Oikos 73, 274–276.
Wright, S.A.I., C.H. Zumoff, L. Schneider, and S.V. Beer. 2001. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 67, 284–292.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bulgari, D., Casati, P., Brusetti, L. et al. Endophytic bacterial diversity in grapevine (Vitis vinifera L.) leaves described by 16S rRNA gene sequence analysis and length heterogeneity-PCR. J Microbiol. 47, 393–401 (2009). https://doi.org/10.1007/s12275-009-0082-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-009-0082-1