Abstract
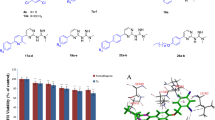
The syntheses of dihydropyrimidinones (DHPMs) using solvent-free grindstone chemistry method. All the synthesized compounds exhibited significant activity against pathogenic bacteria. The current effort has been developed to obtain new DHPM derivatives that focus on the bacterial ribosomal A site RNA as a drug target. Molecular docking simulation analysis was applied to confirm the target specificity of DHPMs. The crystal structure of bacterial 16S rRNA and human 40S rRNA was taken as receptors for docking. Finally, the docking score, binding site interaction analysis revealed that DHPMs exhibit more specificity towards 16S rRNA than known antibiotic amikacin. Accordingly, targeting the bacterial ribosomal A site RNA with potential drug leads promises to overcome the bacterial drug resistance. Even though, anti-neoplastic activities of DHPMs were also predicted through prediction of activity spectra for substances (PASS) tool. Further, the results establish that the DHPMs can serve as perfect leads against bacterial drug resistance.




Similar content being viewed by others
References
Bachi BB (2002) Resistance mechanisms of Gram-positive bacteria. Int J Med Microbiol 292:27–35
Zhou J, Wang G, Zhang L, Ye X (2007) Modifications of aminoglycoside antibiotics targeting RNA. Med Res Rev 27:279–316
Li Y, Shen J, Sun X, Li W, Liu G, Tang Y (2010) Accuracy assessment of protein-based docking programs against RNA targets. J Chem Inf Model 50:1134–1146
Philips A, Milanowska K, Lach G, Bujnicki JM (2013) LigandRNA: computational predictor of RNA-ligand interactions. RNA 19:1605–1616
Ma C, Baker NA, Joseph S, McCammon JA (2002) Binding of aminoglycoside antibiotics to the small ribosomal subunit: a continuum electrostatics investigation. J Am ChemSoc 124:1438–1442
Fullea S, Gohlke H (2009) Molecular recognition of RNA: challenges for modelling interactions and plasticity. J Mol Recognit. doi:10.1002/jmr.1000
Brands M, Endermann R, Gahlmann R, Kruger J, Raddatz S (2003) Dihydropyrimidinones—a new class of anti-staphylococcal antibiotics. Bioorg Med Chem Lett 13:241–245
Kappe CO (1993) 100 years of the biginelli dihydropyrimidine synthesis. Tetrahedron 49(32):6937–6963
Kappe C, Kumar OD, Varma RS (1999) Microwave-assisted high-speed parallel synthesis of 4-aryl-3,4-dihydropyrimidin-2(1H)-ones using a solventless Biginelli condensation protocol. Synthesis 10:1799–803
Stefani HA, Gatti PM (2000) 3,4-Dihydropyrimidin-2(1H)-ones: fast synthesis under microwave irradiation in solvent free conditions. Synth Commun 30:2165–2173
Kappe CO (2000) Recent advances in the Biginelli dihydropyrimidine synthesis. New tricks from an old dog. Acc Chem Res 33(12):879–888
De SK, Gibbs RA (2005) Ruthenium (III) chloride-catalyzed one-pot synthesis of 3,4-dihydropyrimidin-2-(1H)-ones under solvent-free conditions. Synthesis 11:1748–50
Kappe CO, Stadler A (2004) The Biginelli dihydropyrimidine synthesis. Org React 63(1):1–116
Jayakumar S, Shabeer TK (2011) Multicomponent Biginelli synthesis of 3,4-dihydropyrimidin-2(1H)-ones by grindstone technique and evaluation of their biological properties. J Chem Pharm Res 3(6):1089–1096
Deshmukh MB, Prashant AV, Jadhav SD, Mali AR, Jagtap SS, Deshmukh SA (2007) An efficient simple one pot synthesis of dihydropyrimidine-2 (1H) ones using phosphorus pentoxide. Indian J Chem 46B(1545):48
LigPrep, version 2.6. (2012) Schrödinger, LLC, New York, NY
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234
Friensner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, RepaskyMp KEH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid accurate docking and scoring method and assessment of docking accuracy. J Med Chem 47:1739–1749
Vijayalakshmi P, Selvaraj C, Singh SK, Nisha J, Saipriya K, Daisy P (2012) Exploration of the binding of DNA binding ligands to Staphylococcal DNA through QM/MM docking and molecular dynamics simulation. J Biomol Struct Dyn 1–11
Glide, version 5.8. 2012 Schrödinger, LLC, New York, NY
QikProp, version 3.5. 2012 Schrödinger, LLC, New York, NY
Tripathi SK, Selvaraj C, Singh SK, Reddy KK (2012) Molecular docking, QPLD, and ADME prediction studies on HIV-1 integrase leads. Med Chem Res. doi:10.1007/s00044-011-9940-6
Lagunin A, Stepanchikova A, Filimonov D, Poroikov V (2000) PASS: prediction of activity spectra for biologically active substances. Bioinformatics 16:747–748
Poroikov VV, Filimonov DA, Ihlenfeldt WD, Gloriozova TA, Lagunin AA, Borodina YV, Stepanchikova AV, Nicklaus MC (2002) PASS biological activity spectrum predictions in the enhanced open NCI database browser. J Chem Inf Comput Sci 43:228–236
Sadym A, Lagunin A, Filimonov D, Poroikov V (2003) Prediction of biological activity spectra via the internet. SAR QSAR Environ Res 14:339–347
Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ (1999) Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286(5441):971–974
Garcia-Saez I, DeBonis S, Lopez R, Trucco F, Rousseau B, Thuéry P, Kozielski F (2007) Structure of human Eg5 in complex with a new monastrol-based inhibitor bound in the R configuration. J Biol Chem 282(13):9740–9747
Dhumaskar KL, Meena SN, Ghadi SC, Tilve SG (2014) Graphite catalyzed solvent free synthesis of dihydropyrimidin-2(1H)-ones/thiones and their antidiabetic activity. Bioorg Med Chem Lett 24:2897–2899
McCgaughey GB, Gagne M, Rappe AK (1998) π-Stacking interactions. Alive and well in proteins. J BiolChem 273:15458–15463
Dougherty DA (2007) Cation-π interactions involving aromatic amino acids. J Nutr 137:1504S–1508S
Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R (2011) Novel ligands for a purine riboswitch discovered by RNA-ligand docking. Chem Biol 18:324–335
Kadir FA, Kassim NM, Abdulla MA, Yehye WA (2013) PASS-predicted Vitex negundo activity: antioxidant and antiproliferative properties on human hepatoma cells-an in vitro study. BMC Complement Alternat Med 13:343
Kim J, Park C, Ok T, So W, Jo M, Seo M, Kim Y, Sohn JH, Park Y, Ju MK, Kim J, Han SJ, Kim TH, Cechetto J, Nam J, Sommer P, No Z (2012) Discovery of 3,4-dihydropyrimidin-2(1H)-ones with inhibitory activity against HIV-1 replication. Bioorg Med Chem Lett 22(5):2119–2124
Jalali M, Mahdavi M, Memarian HR, Ranjbar M, Soleymani M, Fassihi A, Abedi D (2012) Antimicrobial evaluation of some novel derivatives of 3,4-dihydropyrimidine-2(1H)-one. Res Pharm Sci 7(4):243–247
Törün T, Güngör G, Özmen I, Bölükba Y, Maden E, Bıçakçı B, Ataç G, Sevim T, Tahaoglu K (2005) Side effects associated with the treatment of multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 9(12):1373–1377
Pickering LK, Gearhart P (1979) Effect of time and concentration upon interaction between gentamicin, tobramycin, netilmicin, or amikacin and carbenicillin or ticarcillin. Antimicrob Agents Chemother 15(4):592–596
Acknowledgments
The author SKS thank the Department of Science and Technology (DST) for Fast Track grant (SR/FT/CS-66/2010) and the Department of Biotechnology (DBT), New Delhi, BT/502/NE/TBP/ 2013. One of the authors, SKK, thanks the Conacyt and SEP of Mexican Govt.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ramachandran, V., Arumugasamy, K., Singh, S.K. et al. Synthesis, antibacterial studies, and molecular modeling studies of 3,4-dihydropyrimidinone compounds. J Chem Biol 9, 31–40 (2016). https://doi.org/10.1007/s12154-015-0142-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12154-015-0142-4