Abstract
We investigate the effect that uneven sampling of the environmental gradient has on transfer-function performance using simulated community data. We find that cross-validated estimates of the root mean squared error of prediction can be strongly biased if the observations are very unevenly distributed along the environmental gradient. This biased occurs because species optima are more precisely known (and more analogues are available) in the part of the gradient with most observations, hence estimates are most precise here, and compensate for the less precise estimates in the less well sampled parts of the gradient. We find that weighted averaging and the modern analogue technique are more sensitive to this problem than maximum likelihood, and suggest a way to remove the bias via a segment-wise RMSEP procedure.
Similar content being viewed by others
Introduction
Transfer functions for quantitative reconstructions of environmental variables based on the relationship between species and the environment in a modern training set have been immensely useful tools in the palaeo-sciences. Despite this utility, and the effort spent generating such training sets, there has been little work attempting to optimise the design of training sets. Here we consider the impact of uneven sampling along the environmental gradient.
ter Braak and Looman (1986) demonstrated that the efficiency of weighted averaging (WA) for estimating species’ optima and tolerances approaches that of Gaussian logit regression only when the environmental gradient is evenly sampled. Poorly estimated WA optima are unlikely to give the most reliable reconstructions, so we predict that training sets with evenly sampled gradients should perform better than those with unevenly sampled gradients, and that this difference should be larger with WA than maximum likelihood regression and calibration which uses Gaussian logit regression.
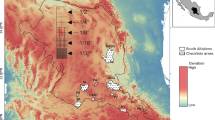
Most training sets are, at least in part, samples-of-convenience and can differ markedly from a uniform distribution in environmental space (Fig. 1). Often, some attempt is made to evenly sample the gradient, but without knowing the magnitude of the performance penalty, this aim is often not prioritised. For some training sets, acquiring a representative set of lakes from a region is prioritised (Dixit et al. 1999); such training sets are unlikely to be evenly sampled along important environmental gradients.
Sampling distributions of real diatom-pH training sets in order of increasing unevenness: a SWAP NW Europe (Birks et al. 1990), b Adirondack USA (Dixit et al. 1993), c Norway (Birks, Boyle and Berge, unpublished), d Sweden (Korsman and Birks 1996), e Finland (Weckström et al. 1997), f NE USA (Dixit et al. 1999). Lines show the segment-wise RMSEP for 1) weighted averaging, 2) maximum likelihood, and 3) modern analogue technique
Ginn et al. (2007) investigated the effect of uneven sampling of the environmental gradient on transfer function performance by taking a large training set and dropping observations from the more densely sampled parts of the gradient until the remaining observations were approximately evenly distributed along the gradient. Surprisingly, they found that the cross-validation performance statistics from the full data set and the uniform data set were similar.
We adopt an alternative strategy, using simulated community data to develop training sets for unevenly sampled environmental gradients, and testing the performance of different transfer function procedures by both cross-validation and with an evenly sampled independent test set.
Methods
Minchin (1987) introduced a method for simulating realistic looking community patterns along environmental gradients using generalised beta distributions to represent species response curves. We implement his method in the statistical language R version 2.11.1 (R Development Core Team 2010) to generate species distributions and simulated assemblages along environmental gradients.
We generated species response curves on three orthogonal environmental gradients, which should approximate the dimensionality of many data sets. The gradient which we hope to reconstruct was 100 units long; two secondary nuisance variables were 60 units long. Species optima for thirty simulated species were uniformly distributed along the environmental gradients, with their maximum abundances drawn from a uniform distribution. Both shape parameters of the beta distributions were set to four, which produces symmetrical, near-Gaussian responses. The range or niche width of each species was set to 200 units. From these response curves, counts of 300 individuals were simulated and relative abundances calculated.
Six diatom-pH training sets, with between 96 and 241 observations, were chosen to represent different degrees of unevenness along the gradient of interest (Fig. 1). For each of these six diatom-pH training-sets, we generated two simulated training sets, one that matches the distribution of sites along the pH gradient, rescaled to fill the range 0–100, and a second that contains as many observations, evenly distributed over the range 0–100. We also generated an independent test-set with 100 evenly distributed observations. For all data sets, the secondary gradients were uniformly sampled.
The length of the first detrended correspondence axis of the simulated data is about 3 SD units of compositions turnover. This is comparable with many diatom training sets (Korsman and Birks 1996; ter Braak and Juggins 1993).
The unevenness of the sample was quantified as the standard deviation of the number of sites in each tenth of the gradient. When comparing training sets with a different number of sites, this value was divided by the total number of sites.
Transfer functions were generated for each training set using weighted averaging with inverse deshrinking (WA; Birks et al. 1990); maximum likelihood regression and calibration (ML; ter Braak and Looman 1986); and the modern analogue technique (MAT; Prell 1985) using squared chord distances. We calculated MAT with three analogues as in a trial run this gave the best performance with the independent test set. Transfer functions were run using the rioja library version 0.5-6 (Juggins 2009). The performance of the training sets was assessed by the root mean square error of prediction (RMSEP), the correlation between the predicted and “observed” environmental variables (r²), and the absolute value of the maximum bias. Maximum bias was calculated by dividing the environmental gradient into ten equally spaced segments, calculating the mean of the residual for each segment, and taking the largest of these ten values. Maximum bias quantifies the tendency for the model to over- or under-estimate somewhere along the gradient (ter Braak and Juggins 1993). Performance was measured for both bootstrap (with 500 bootstrap replicates) and leave-one-out cross-validation, and with the independent test set. The performance of the unevenly sampled simulated training-sets is expressed relative to the performance of the evenly sampled training-sets with the same number of observations. This standardises the results, so different training sets can be compared. The results presented are the mean of 50 trials with different simulated species configurations.
To investigate the relationship between bias in the transfer function performance statistics and the unevenness of the distribution of observations along the environmental gradient in more detail, we took an unevenly sampled gradient and redistributed observations from over-sampled parts of the gradient to under-sampled parts of the gradient. We did this with the NE USA pH distribution, dividing the gradient into ten equal segments and deleting (adding) observations in segments where there are excess (insufficient) observations. For each trial, we added/deleted between 0 and 100% of the excess/insufficiency in each segment in 5% increments. For each of the generated gradients, we simulated assemblage data and estimated the RMSEP both by cross-validation and for an independent test set. We report the ratio of these RMSEPs. Each trial was repeated ten times with different simulated species configurations.
Results
Simulated assemblages data with the distribution of observation along the environmental gradient matching the six diatom-pH training sets, have, in most cases, similar cross-validation RMSEP to training sets with as many observations evenly distributed along the environmental gradient, regardless of transfer function method. This is shown (Fig. 2) by the relative RMSEP being close to one. Only training sets with the most extreme unevenness have a median relative RMSEP significantly above one. For WA and MAT, the RMSEP of the evenly sampled independent test set predicted with the unevenly sampled training set is worse than the cross-validation result would predict. This deterioration is most marked for the most unevenly sampled training sets and for MAT. With ML the performance of the test set and the cross-validation result are, for most cases, similar.
Boxplot of cross-validation (white) and test set (grey) RMSEPs for different simulated training sets with the same distribution of observations along the environmental gradient as the original diatom-pH training set. The upper panel shows WA results, the middle panel ML results and the lower panel MAT results. RMSEPs are standardised relative to that of an evenly sampled training set with the same number of observations. The results are from 50 trials. Large values indicate worse performance
The cross-validation r² between the predicted and “observed” environmental variables for the three most even training sets is similar to the cross-validation performance of the evenly sampled training set performance (Fig. 3), so the relative r² was near one. The r² for the three most unevenly sampled training sets is markedly worse. Even when calculated with the most uneven training sets, the r² for the independent test set predicted with ML is only slightly worse than when calculated using an evenly sampled training set. In contrast, the r² of the independent test-set predicted with WA or MAT is worse for the most uneven training sets than with the evenly sampled training set.
Boxplot of cross-validation (white) and independent test set (grey) r² for different simulated training sets with the same distribution of observations along the environmental gradient as the original diatom-pH training set. The upper panel shows WA results, the middle panel ML results and the lower panel MAT results. The r² values are standardised relative to that of an evenly sampled training set with the same number of observations. The results are from 50 trials. Small values indicate worse performance
The cross-validation maximum bias is larger for the unevenly distributed data sets than for the evenly distributed data sets (relative maximum bias >1) (Fig. 4). This difference is most marked for the most unevenly distributed training sets. The performance of the evenly sampled independent test set is, in most cases, similar to the cross-validation results, but for the most unevenly sampled training sets, it is better. The larger maximum bias for the most uneven training sets under cross-validation rather than for the evenly sampled independent test set is probably because of the low number of observations in some segments of the gradient which will make this metric prone to noise.
Boxplot of cross-validation (white) and independent test set (grey) maximum bias for different simulated training sets with the same distribution of observations along the environmental gradient as the original diatom-pH training set. The upper panel shows WA results, the middle panel ML results and the lower panel MAT results. The maximum bias values are standardised relative to that of an evenly sampled training set with the same number of observations. The results are from 50 trials. Large values indicate worse performance
Bootstrap cross-validation results are essentially identical to the LOO results and are not shown.
Discussion
The effect of uneven sampling of the environmental gradient on species optima, predicted by ter Braak and Looman (1986), has been noted, for example, by Cameron et al. (1999) who ascribed differences between the WA optima for taxa in the SWAP and AL:PE training sets to differences in the distribution of sites in the training set. The impact of uneven sampling on performance statistics of transfer functions has not previously been fully explored. Ginn et al. (2007) found no benefit from even sampling on the cross-validation performance (for one transfer function method they found the r2 to be marginally higher with the evenly distributed training set, but the RMSEP was worse for all methods).
This result was contrary to what Ginn et al. (2007) expected, but is explicable following our results. The cross-validation RMSEP for their unevenly sampled training set is biased, being lower than would be expected for an evenly distributed independent test-set. The cross-validation RMSEP of their evenly sampled training set is unbiased, and therefore fails to outperform the unevenly sampled training set. Interpretation of the results of Ginn et al. (2007) is complicated by the different number of sites in the full and the evenly sampled training sets.
The leave-one-out cross-validation RMSEP is biased because the part of the gradient with most observations is the part where the species optima are most precisely known (or where most available potential analogues are) and hence the part of the gradient where estimates are most reliable. This compensates for the greater uncertainty in the few observations in the less densely sampled parts of the gradient, whereas the evenly sampled independent test set tests all parts of the gradient equally. This bias implies that RMSEP estimated for unequally sampled gradients may be over-optimistic.
The cross-validation r² is lower for the most unevenly sampled training sets than the evenly sampled training set because many of the observation are in a restricted part of the range so their variance is poorly explained by the predictions. The independent test set has a lower r² when predicted with the unevenly sampled training set because the species optima from the under-sampled part of the gradient are less well estimated.
ML is less affected by uneven sampling than WA. Following the finding of ter Braak and Looman (1986) that ML is more efficient at estimating optima along unevenly sampled gradients, this is not surprising. However, test set performance with ML is not consistently better than with WA. This may be because ML is sensitive to over-dispersion in the species data (Telford et al. unpublished data). MAT has the greatest problems with an unevenly sampled gradient. Observations in the poorly sampled parts of the gradient lack sufficient good analogues.
For the training sets generated here, the RMSEP of the evenly sampled independent test set is up to 40% larger than the LOO cross-validated RMSEP with WA. This is potentially large with respect to differences in performance between models (Telford et al. unpublished data) and some other sources of performance bias (Telford et al. 2004). Since generating a completely evenly distributed training set would be very difficult in many cases, it is useful to know what the sensitivity to increasing unevenness is so as to provide a more realistic target. Figure 5 shows how bias changes for the NE USA training set distribution as an increasing number of observations are moved from the over-sampled to the under-sampled segments of the gradient, reducing the standard deviation of the number of observations in each tenth of the gradient. For this 241 observation training set, a standard deviation of ten sites per tenth of the gradient (which would have a mean of 24 observations) is only slightly worse than the completely even case. For real data, the degree of unevenness that can occur before the performance statistics become greatly biased will be dependent on training-set size, noise level and on the specifics of the species’ niche widths, but it suggests that training sets more even than the SWAP data set should have only a small performance bias.
Ratio of independent test set RMSEP to cross-validated RMSEP against unevenness for the NE USA distribution, beginning with the original distribution, and then making it more even by redistributing observations from over- to under-sampled portions of the gradient. cv cross-validation, sd standard deviation
Since it will sometimes be impossible to collect a sufficiently even sample to be confident that the bias in cross-validation RMSEP is minimal, it is useful to be able to correct for the bias. Figure 6 shows the segment-wise RMSEP calculated for simulated data by both cross-validation and for an independent test set. The two results are very similar. This suggests than an unbiased estimate of the RMSEP can be calculated by combining the segment-wise RMSEPs. This can be done by taking the root mean square of the segment-wise RMSEPs. Table 1 shows the RMSEP and the segment-wise RMSEP, calculated using 10 equal-sized segments, for the six diatom-pH training sets. Segment-wise RMSEP is only slightly higher than RMSEP for the most evenly distributed training sets, but much higher for the most uneven. The difference between the RMSEP and the segment-wise RMSEP is highly correlated with the absolute value maximum bias (r² > 0.9) for all three methods.
The under-estimation of the RMSEP by cross-validation for unevenly sampled gradients will only be a problem when trying to reconstruct the under-sampled parts of the gradient. For reconstructions from the over-sampled parts of the gradient, RMSEP may even be pessimistic.
Conclusions
Cross-validation underestimates the uncertainty in unevenly sampled gradients, although some unevenness is possible before the bias in RMSEP becomes large. Maximum likelihood is the most robust method, the modern analogue technique it the least robust. Calculating the RMSEP by segments can correct for this bias.
References
Birks HJB, Line JM, Juggins S, Stevenson AC, ter Braak CJF (1990) Diatoms and pH reconstruction. Philoso Trans R Soc B-Biol Sci 327:263–278
Cameron NG, Birks HJB, Jones VJ, Berges F, Catalan J, Flower RJ, Garcia J, Kawecka B, Koinig KA, Marchetto A, Sánchez-Castillo P, Schmidt R, Šiško M, Solovieva N, Šefková E, Toro M (1999) Surface-sediment and epilithic diatom pH calibration sets for remote European mountain lakes (AL:PE Project) and their comparison with the Surface Waters Acidification Programme (SWAP) calibration set. J Paleolimnol 22:291–317
Dixit SS, Cumming BF, Birks HJB, Smol JP, Kingston JC, Uutala AJ, Charles DF, Camburn KE (1993) Diatom assemblages from Adirondack lakes (New York, USA) and the development of inference models for retrospective environmental assessment. J Paleolimnol 8:27–47
Dixit SS, Smol JP, Charles DF, Hughes RM, Paulsen SG, Collins GB (1999) Assessing water quality changes in the lakes of the northeastern United States using sediment diatoms. Can J Fish Aquat Sci 56:131–152
Ginn BK, Cumming BF, Smol JP (2007) Diatom-based environmental inferences and model comparisons from 494 northeastern North American lakes. J Phycol 43:647–661
Juggins S (2009) Rioja: an R package for the analysis of quaternary science data, version 0.5-4
Korsman T, Birks HJB (1996) Diatom-based reconstruction from northern Sweden: a comparison of reconstruction techniques. J Paleolimnol 15:65–77
Minchin PR (1987) Simulation of multidimensional community patterns: towards a comprehensive model. Vegetatio 71:145–156
Prell WL (1985) The stability of low-latitude sea-surface temperatures: an evaluation of the CLIMAP reconstruction with emphasis on the positive SST anomalies. Department of Energy, Washington, 60 pp
R Development Core Team (2010) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Telford RJ, Andersson C, Birks HJB, Juggins S (2004) Biases in the estimation of transfer function prediction errors. Paleoceanography 19: Artn Pa4014
ter Braak CJF, Juggins S (1993) Weighted averaging partial least-squares regression (WA-PLS)—an improved method for reconstructing environmental variables from species assemblages. Hydrobiologia 269:485–502
ter Braak CJF, Looman CWN (1986) Weighted averaging, logistic-regression and the Gaussian response model. Vegetatio 65:3–11
Weckström J, Korhola A, Blom T (1997) The relationship between diatoms and water temperature in thirty subarctic Fennoscandian Lakes. Arctic Alpine Res 29:75–92
Acknowledgments
This work was supported by Norwegian Research Council projects ARCTREC and PES. This is publication no. A319 from the Bjerknes Centre for Climate Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Telford, R.J., Birks, H.J.B. Effect of uneven sampling along an environmental gradient on transfer-function performance. J Paleolimnol 46, 99–106 (2011). https://doi.org/10.1007/s10933-011-9523-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10933-011-9523-z