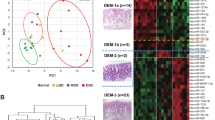
Abstract
Background
Current strategies fail to identify most patients with esophageal adenocarcinoma (EAC) before the disease becomes advanced and incurable. Given the dismal prognosis associated with EAC, improvements in detection of early-stage esophageal neoplasia are needed.
Aim
We sought to assess whether differential expression of microRNAs could discriminate between squamous epithelium, Barrett’s esophagus (BE), and EAC.
Methods
We analyzed microRNA expression in a discovery cohort of human endoscopic biopsy samples from 36 patients representing normal squamous esophagus (n = 11), BE (n = 14), and high-grade dysplasia/EAC (n = 11). RNA was assessed using microarrays representing 847 human microRNAs followed by quantitative real-time polymerase chain reaction (qRT-PCR) verification of nine microRNAs. In a second cohort (n = 18), qRT-PCR validation of five miRNAs was performed. Expression of 59 microRNAs associated with BE/EAC in the literature was assessed in our training cohort. Known esophageal cell lines were used to compare miRNA expression to tissue miRNAs.
Results
After controlling for multiple comparisons, we found 34 miRNAs differentially expressed between squamous esophagus and BE/EAC by microarray analysis. However, miRNA expression did not reliably differentiate non-dysplastic BE from EAC. In the validation cohort, all five microRNAs selected for qRT-PCR validation differentiated between squamous samples and BE/EAC. Microarray results supported 14 of the previously reported microRNAs associated with BE/EAC in the literature. Cell lines did not generally reflect miRNA expression found in vivo.
Conclusions
These data indicate that miRNAs differ between squamous esophageal epithelium and BE/EAC, but do not distinguish between BE and EAC. We suggest prospective evaluation of miRNAs in patients at high risk for EAC.


Similar content being viewed by others
References
Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett’s esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122:26–33.
Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011;365:1375–1383.
Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association medical position statement on the management of Barrett’s esophagus. Gastroenterology. 2011;140:1084–1091.
National Cancer Institute. SEER Cancer Statistics Review, 2011. Available at: http://seer.cancer.gov/csr/1975_2008/; 2012. Accessed 14 March 2012.
Rubenstein JH, Mattek N, Eisen G. Age- and sex-specific yield of Barrett’s esophagus by endoscopy indication. Gastrointest Endosc. 2010;71:21–27.
Shaheen NJ, Crosby MA, Bozymski EM, Sandler RS. Is there publication bias in the reporting of cancer risk in Barrett’s esophagus? Gastroenterology. 2000;119:333–338.
Ben-Menachem T, Decker GA, Early DS, et al. Adverse events of upper GI endoscopy. Gastrointest Endosc. 2012;76:707–718.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838.
Croce CM. miRNAs in the spotlight: understanding cancer gene dependency. Nat Med. 2011;17:935–936.
Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529.
Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53.
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261.
Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association technical review on the management of Barrett’s esophagus. Gastroenterology. 2011;140:e18–e52. (quiz e13).
Opitz L, Salinas-Riester G, Grade M, et al. Impact of RNA degradation on gene expression profiling. BMC Med Genomics. 2010;3:36.
R_Core_Team. R: A Language and Environment for Statistical Computing. New York: R Foundation for Statistical Computing; 2012.
Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.
Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315.
Koshiol J, Wang E, Zhao Y, Marincola F, Landi MT. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol Biomarkers Prev. 2010;19:907–911.
Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264.
Chen DT, Hernandez JM, Shibata D, et al. Complementary strand microRNAs mediate acquisition of metastatic potential in colonic adenocarcinoma. J Gastrointest Surg. 2012;16:905–912. (discussion 912–913).
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144.
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158.
Kendall M. A new measure of rank correlation. Biometrika. 1938;30:81–89.
Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. (discussion 260).
Yang H, Gu J, Wang KK, et al. MicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–5752.
Bansal A, Lee IH, Hong X, et al. Feasibility of MicroRNAs as biomarkers for Barrett’s esophagus progression: a pilot cross-sectional, phase 2 biomarker study. Am J Gastroenterol. 2011;106:1055–1063.
Leidner RS, Ravi L, Leahy P, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett’s esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473–479.
Fassan M, Volinia S, Palatini J, et al. MicroRNA expression profiling in human Barrett’s carcinogenesis. Int J Cancer. 2011;129:1661–1670.
Mathe EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–6200.
Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853–861.
Huo X, Zhang HY, Zhang XI, et al. Acid and bile salt-induced CDX2 expression differs in esophageal squamous cells from patients with and without Barrett’s esophagus. Gastroenterology. 2010;139(194–203):e191.
Zhang HY, Zhang X, Chen X, et al. Differences in activity and phosphorylation of MAPK enzymes in esophageal squamous cells of GERD patients with and without Barrett’s esophagus. Am J Physiol Gastrointest Liver Physiol. 2008;295:G470–G478.
Jaiswal KR, Morales CP, Feagins LA, et al. Characterization of telomerase-immortalized, non-neoplastic, human Barrett’s cell line (BAR-T). Dis Esophagus. 2007;20:256–264.
Hormi-Carver K, Zhang X, Zhang HY, et al. Unlike esophageal squamous cells, Barrett’s epithelial cells resist apoptosis by activating the nuclear factor-kappaB pathway. Cancer Res. 2009;69:672–677.
Jonckheere A. A distribution-free k-sample test again ordered alternatives. Biometrika. 1954;41:133–145.
Terpstra T. The asymptotic normality and consistency of Kendall’s test against trend, when ties are present in one ranking. Indag Math. 1952;14:327–333.
Clinfun: Clinical Trial Design and Data Analysis Functions. R Package version 1.0.3. Available at: http://CRAN.R-project.org/package=clinfun; 2012. Accessed 08 November 2012.
Sarkar D. Lattice: Multivariate Data Visualization with R. New York: Springer; 2008.
Wu X, Ajani JA, Gu J, et al. MicroRNA expression signatures during malignant progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Prev Res (Phila). 2013;6:196–205.
McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664.
van Baal JW, Verbeek RE, Bus P, et al. microRNA-145 in Barrett’s oesophagus: regulating BMP4 signalling via GATA6. Gut. 2010;62:664–675.
Kadri SR, Lao-Sirieix P, O’Donovan M, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett’s oesophagus in primary care: cohort study. BMJ. 2010;341:c4372.
Montgomery E, Bronner MP, Goldblum JR. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368–378.
Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98.
Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379.
Fassan M, Pizzi M, Battaglia G, et al. Programmed cell death 4 (PDCD4) expression during multistep Barrett’s carcinogenesis. J Clin Pathol. 2010;63:692–696.
Acknowledgments
This work was supported by an American College of Gastroenterology Junior Faculty Development Award (KG) and by the Emeline Brown Cancer Genomics Research Fund (KG). Support was also from the National Cancer Institute U54CA163060 (NS).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Garman, K.S., Owzar, K., Hauser, E.R. et al. MicroRNA Expression Differentiates Squamous Epithelium from Barrett’s Esophagus and Esophageal Cancer. Dig Dis Sci 58, 3178–3188 (2013). https://doi.org/10.1007/s10620-013-2806-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-013-2806-7