Abstract
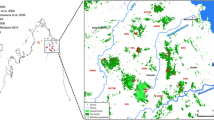
Microcebus ravelobensis is an endangered nocturnal primate endemic to northwestern Madagascar. This part of the island is subject to extensive human intervention leading to massive habitat destruction and fragmentation. We investigated the degree of genetic differentiation among remaining populations using mitochondrial control region sequences (479–482 bases). Nine populations were sampled from the hypothesized geographic range. The region is composed of three inter-river systems (IRSs). Samples were collected in three areas of continuous forests (CFs) and six isolated forest fragments (IFFs) of different sizes. We identified 27 haplotypes in 114 animals, with CFs and IFFs harbouring 5–6 and 1–3 haplotypes, respectively. All IFFs were significantly differentiated from each other with high ΦST values and sets of unique haplotypes. The rivers constitute significant dispersal barriers with over 82% of the molecular variation being attributed to the divergence among the IRSs. The data suggest a deep and so far unknown split within the rufous mouse lemurs of northwestern Madagascar. The limited data base and the lack of ecological and morphological data do not allow definite taxonomic classification at this stage. However, the results clearly indicate that M. ravelobensis consists of three evolutionary significant units, possibly cryptic species, which warrant urgent and separate conservation efforts.



Similar content being viewed by others
References
Avise JC (2000) Phylogeography. Harvard University Press, Cambridge, MA
Barrett EM, Gurnell J, Malarky G, Deaville R, Bruford MW (1999) Genetic structure of fragmented populations of red squirrel (Sciurus vulgaris) in the UK. Mol Ecol 8:S55–S63
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Dewar RE (1997) Were people responsible for the extinction of Madagascar’s subfossils, and how will we ever know? In: Goodman SM, Petterson BD (eds.) Natural change and human impact in Madagascar. Smithsonian Institution Press, Washington, pp. 364–377
Frazer DF, Bernatchez L (2001) Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10:2741–2752
Fredsted T, Pertoldi C, Olesen JM, Eberle M, Kappeler PM (2004) Microgeographic heterogeneity in spatial distribution and mtDNA variability of gray mouse lemurs (Microcebus murinus, Primates: Cheirogaleidae). Behav Ecol Sociobiol 56:393–403
Gerloff U, Hartung B, Fruth B, Hohmann G, Tautz D (1999) Intracommunity relationships, dispersal pattern and paternity success in a wild living community of Bonobos (Pan paniscus) determined from DNA analysis of faecal samples. Proc R Soc Lond B 266:1189–1195
Hofreiter M, Siedel H, Van Neer W, Vigilant L (2003) Mitochondrial DNA sequence from an enigmatic gorilla population (Gorilla gorilla uellensis). Am J Phys Anthropol 121:361–368
Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612
Johnson JA, Bellinger MR, Toepfer JE, Dunn P (2004) Temporal changes in allele frequencies and low effective population size in greater prairie-chickens. Mol Ecol 13:2617–2630
Kappeler PM, Rasoloarison RM, Razafimanantsoa L, Walter L, Roos C (2005) Morphology, behaviour and molecular evolution of giant mouse lemurs (Mirza spp.) gray, 1970, with description of a new species. Prim Report 71:3–26
Louis EE, Coles ME, Andrintompohavana R, Sommer JA, Engberg SE, Zaonarivelo JR, Mayor MI, Brenneman RA (2006) Revision of the Mouse Lemurs (Microcebus) of Eastern Madagascar. Int J Primotol 27:347–389
Moritz C (1994) Defining “evolutionary significant units” for conservation. Trends Ecol Evol 9:373–375
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Pastorini J, Martin RD, Ehresmann P, Zimmermann E, Forstner MR (2001) Molecular phylogeny of the lemur family Cheirogaleidae (primates) based on mitochondrial DNA sequences. Mol Phylogenet Evol 19:45–56
Pastorini J, Thalmann U, Martin RD (2003) A molecular approach to comparative phylogeography of extant Malagasy lemurs. Proc Natl Acad Sci USA 100:5879–5884
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Radespiel U, Lutermann H, Schmelting B, Bruford MW, Zimmermann E (2003) Patterns and dynamics of sex-biased dispersal in a nocturnal primate, the grey mouse lemur, Microcebus murinus. Anim Behav 65:709–719
Radespiel U, Raveloson H (2001) Preliminary study on the lemur communities at three sites of dry deciduous forest in the Réserve Naturelle d’Ankarafantsika. Lemur News 6:22–24
Radespiel U, Sarikaya Z, Zimmermann E (2001) Sociogenetic structure in a free-living nocturnal primate population: sex-specific differences in the grey mouse lemur (Microcebus murinus). Behav Ecol Sociobiol 50:493–502
Randrianambinina B, Rasoloharijaona S, Rakotosamimanana B (2003) Inventaires des communautés lémuriennes dans la réserve spéciale de Bora au nord-ouest et la forêt domaniale de Mahilaka-Maromandia au nord de Madagascar. Lemur News 8:15–18
Rakotoarisoa J-A (1997) A cultural history of Madagascar: evolution and interpretation of the archeological evidence. In: Goodman SM, Petterson BD (eds.) Natural change and human impact in Madagascar. Smithsonian Institution Press, Washington, pp. 331–341
Rasoloarison RM, Goodman SM, Ganzhorn JU (2000) Taxonomic revision of mouse lemurs (Microcebus) in the western portions of Madagascar. Int J Primotol 21:963–1019
Rendigs A, Radespiel U, Wrogemann D, Zimmermann E (2003) Relationship between microhabitat structure and distribution of mouse lemurs (Microcebus spp.) in Northwestern Madagascar. Int J Primatol 24:47–64
Rozas J, Sanchez-DelBarrio JC, Messenguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.000: a software for population genetics data analysis
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90
Smith AP (1997) Deforestation, fragmentation, and reserve design in western Madagascar. In: Laurance WF, Bierregard RO Jr (eds.) Tropical forest remnants: ecology, management, and conservation of fragmented communities. University of Chicago Press, Chicago, pp. 415–441
Swofford DL (1998) PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA
Tallman DA, Draheim HM, Mills LS, Allendorf FW (2002) Insights into recently fragmented vole populations from combined genetic and demographic data. Mol Ecol 11:699–709
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thalmann O, Hebler J, Poinar HN, Paabo S, Vigilant L (2004) Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol Ecol 13:321–335
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Weidt A, Hagenah N, Randrianambinina B, Radespiel U, Zimmermann E (2004) Social organization of the golden brown mouse lemur (Microcebus ravelobensis). Am J Phys Anthropol 123:40–51
Wimmer B, Tautz D, Kappeler PM (2002) The genetic population structure of the gray mouse lemur (Microcebus murinus), a basal primate from Madagascar. Behav Ecol Sociobiol 52:166–175
Wright HT, Rakotoarisoa J-A (1997) Cultural transformations and the impacts on the environments of Madagascar. In: Goodman SM, Petterson BD (eds.) Natural change and human impact in Madagascar. Smithsonian Institution Press, Washington, pp. 309–330
Yoder AD, Cartmill M, Ruvolo M, Smith K, Vilgalys R (1996) Ancient single origin for Malagasy primates. Proc Natl Acad Sci USA 93:5122–5126
Yoder AD, Rasoloarison RM, Goodman SM, Irwin JA, Atsalis S, Ravosa MJ, Ganzhorn JU (2000) Remarkable species diversity in Malagasy mouse lemurs (primates, Microcebus). Proc Natl Acad Sci USA 97:11325–11330
Yoder AD, Yang Z (2000) Estimation of primate speciation dates using local molecular clocks. Mol Biol Evol 17:1081–1090
Yoder AD, Yang Z (2004) Divergence dates for Malagasy lemurs estimated from multiple gene loci: geological and evolutionary context. Mol Ecol 13:757–773
Zimmermann E, Cepok S, Rakotoarison N, Zietemann V, Radespiel U (1998) Sympatric mouse lemurs in north-west Madagascar: a new rufous mouse lemur species (Microcebus ravelobensis). Folia Primatol 69:106–114
Acknowledgements
We would like to thank the following institutions and persons for their support and the permissions to conduct the necessary fieldwork in Madagascar: Le Ministère des Eaux et Forêts, Chantal Andrianarivo (ANGAP), University of Antananarivo (Olga Ramilijaona and Daniel Rakotondravony), and the late Berthe Rakotosamimanana. For help and assistance in the field, we thank Mathias Craul, Jean-Aimé Rakotonirina, Todisoa Razafimamonjy. Many thanks go to all students and colleagues who helped collecting samples over the course of the last 4 years and especially to Pia Braune, Nicole Hagenah, Kathrin Marquart, Romule Rakotondravony, Blanchard Randrianambinina, Solofo Rasoloharijaona, and Andrea Weidt. We are very grateful to Elke Zimmermann for her constant support during the course of the work. We thank Lounès Chikhi and Mike Bruford for the helpful comments on the analyses and the manuscript. K.G. thanks Heike Pröhl for valuable discussions. Thanks go to the Institute of Zoology, Zoological Society of London, UK, for providing molecular facilities for part of the study. The manuscript was greatly improved by the comments of two anonymous reviewers. The study was financially supported by DFG (Ra 502/7-1) and Conservation International. We also gratefully acknowledge the Scholarship of the German National Academic Foundation for the financial support of K.G. throughout the study and especially during the fieldwork in Madagascar.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Summary list of samples used in this study. The source population, the respective IRS, year of sampling, sex and haplotype assignment as well as the sampler of each individual are provided. The sample ID is composed of the individual number, followed by the year of sampling and by an abbreviation for the population. All sequences have been deposited in GenBank under the Accession numbers EF065272–EF065385.
Sample ID | Population | IRS | Year | Sex | Haplotype | Samplera |
|---|---|---|---|---|---|---|
012y00jbb | JBB | I | 2000 | m | H11 | UR, P. Braune |
014y00jbb | JBB | I | 2000 | f | H13 | UR, P. Braune |
008y00jbb | JBB | I | 2000 | f | H15 | UR, P. Braune |
011y00jbb | JBB | I | 2000 | f | H18 | UR, P. Braune |
005y00jbb | JBB | I | 2000 | m | H11 | UR, P. Braune |
016y00jbb | JBB | I | 2000 | f | H15 | UR, P. Braune |
021y00jbb | JBB | I | 2000 | m | H11 | UR, P. Braune |
007y00jbb | JBB | I | 2000 | f | H21 | UR, P. Braune |
006y00jbb | JBB | I | 2000 | m | H11 | UR, P. Braune |
002y00jbb | JBB | I | 2000 | m | H18 | UR, P. Braune |
004y00jbb | JBB | I | 2000 | f | H18 | UR, P. Braune |
010y00jbb | JBB | I | 2000 | f | H18 | UR, P. Braune |
013y00jbb | JBB | I | 2000 | m | H11 | UR, P. Braune |
015y00jbb | JBB | I | 2000 | f | H11 | UR, P. Braune |
078y00bev | Bevazaha | I | 2003 | m | H16 | R. Rakotondravony |
095y00bev | Bevazaha | I | 2003 | m | H16 | R. Rakotondravony |
001y00bev | Bevazaha | I | 2000 | f | H12 | UR |
089y03bev | Bevazaha | I | 2000 | f | H12 | UR |
098y03bev | Bevazaha | I | 2003 | f | H13 | R. Rakotondravony |
088y03bev | Bevazaha | I | 2003 | m | H16 | R. Rakotondravony |
084y03bev | Bevazaha | I | 2003 | m | H16 | R. Rakotondravony |
085y03bev | Bevazaha | I | 2003 | f | H16 | R. Rakotondravony |
079y03bev | Bevazaha | I | 2003 | f | H16 | R. Rakotondravony |
116y03bev | Bevazaha | I | 2003 | f | H23 | R. Rakotondravony |
005y00bev | Bevazaha | I | 2000 | m | H12 | UR |
006y00bev | Bevazaha | I | 2000 | f | H12 | UR |
080y03bev | Bevazaha | I | 2003 | m | H22 | R. Rakotondravony |
003y00bev | Bevazaha | I | 2000 | f | H17 | UR |
004y00bev | Bevazaha | I | 2000 | m | H17 | UR |
002y00bev | Bevazaha | I | 2000 | f | H17 | UR |
022y01stm | Ste.Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
028y01stm | Ste.Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
012y01stm | Ste.Marie | I | 2001 | m | H24 | UR, KG, K. Marquart |
020y01stm | Ste.Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
003y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
006y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
011y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
021y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
016y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
027y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
005y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
004y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
002y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
014y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
013y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
019y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
008y01stm | Ste. Marie | I | 2001 | f | H14 | UR, KG, K. Marquart |
018y01stm | Ste. Marie | I | 2001 | m | H14 | UR, KG, K. Marquart |
103y03mari | Mariarano | I | 2003 | f | H20 | GO |
106y03mari | Mariarano | I | 2003 | m | H10 | GO |
061y03mari | Mariarano | I | 2003 | f | H10 | GO |
096y03mari | Mariarano | I | 2003 | f | H10 | GO |
081y03mari | Mariarano | I | 2003 | f | H20 | GO |
057y03mari | Mariarano | I | 2003 | f | H25 | GO |
097y03mari | Mariarano | I | 2003 | f | H10 | GO |
079y03mari | Mariarano | I | 2003 | f | H20 | GO |
085y03mari | Mariarano | I | 2003 | f | H10 | GO |
075y03mari | Mariarano | I | 2003 | m | H20 | GO |
058y03mari | Mariarano | I | 2003 | m | H20 | GO |
101y03mari | Mariarano | I | 2003 | f | H10 | GO |
099y03mari | Mariarano | I | 2003 | f | H10 | GO |
051y03mari | Mariarano | I | 2003 | f | H20 | GO |
086y03mari | Mariarano | I | 2003 | f | H10 | GO |
367y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
359y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
368y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
358y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
369y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
365y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
360y03tsia | Tsiaramaso | I | 2003 | m | H19 | GO, KG |
371y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
370y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
364y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
363y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
366y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
362y03tsia | Tsiaramaso | I | 2003 | m | H19 | GO, KG |
361y03tsia | Tsiaramaso | I | 2003 | m | H19 | GO, KG |
372y03tsia | Tsiaramaso | I | 2003 | f | H19 | GO, KG |
340y03ata | Maroakata | II | 2003 | m | H5 | GO, KG |
311y03ata | Maroakata | II | 2003 | m | H5 | GO, KG |
308y03ata | Maroakata | II | 2003 | f | H6 | GO, KG |
323y03ata | Maroakata | II | 2003 | f | H6 | GO, KG |
345y03ata | Maroakata | II | 2003 | f | H26 | GO, KG |
332y03ata | Maroakata | II | 2003 | m | H5 | GO, KG |
312y03ata | Maroakata | II | 2003 | m | H5 | GO, KG |
314y03ata | Maroakata | II | 2003 | f | H5 | GO, KG |
326y03ata | Maroakata | II | 2003 | f | H5 | GO, KG |
327y03ata | Maroakata | II | 2003 | f | H5 | GO, KG |
301y03ata | Maroakata | II | 2003 | f | H7 | GO, KG |
313y03ata | Maroakata | II | 2003 | m | H7 | GO, KG |
347y03ata | Maroakata | II | 2003 | f | H8 | GO, KG |
317y03ata | Maroakata | II | 2003 | m | H8 | GO, KG |
339y03ata | Maroakata | II | 2003 | f | H5 | GO, KG |
307y03ata | Maroakata | II | 2003 | f | H6 | GO, KG |
318y03ata | Maroakata | II | 2003 | f | H8 | GO, KG |
319y03ata | Maroakata | II | 2003 | f | H8 | GO, KG |
151y03est | Mahajamba-Est | II | 2003 | m | H9 | UR, R. Rakotondravony |
154y03est | Mahajamba-Est | II | 2003 | f | H9 | UR, R. Rakotondravony |
153y03est | Mahajamba-Est | II | 2003 | f | H9 | UR, R. Rakotondravony |
149y03est | Mahajamba-Est | II | 2003 | f | H9 | UR, R. Rakotondravony |
152y03est | Mahajamba-Est | II | 2003 | f | H9 | UR, R. Rakotondravony |
148y03est | Mahajamba-Est | II | 2003 | f | H9 | UR, R. Rakotondravony |
357y03amb | Ambongabe | III | 2003 | f | H2 | GO, KG |
356y03amb | Ambongabe | III | 2003 | f | H2 | GO, KG |
349y03amb | Ambongabe | III | 2003 | m | H2 | GO, KG |
352y03amb | Ambongabe | III | 2003 | m | H2 | GO, KG |
355y03amb | Ambongabe | III | 2003 | f | H1 | GO, KG |
348y03amb | Ambongabe | III | 2003 | f | H3 | GO, KG |
353y03amb | Ambongabe | III | 2003 | f | H3 | GO, KG |
354y03amb | Ambongabe | III | 2003 | m | H3 | GO, KG |
350y03amb | Ambongabe | III | 2003 | f | H1 | GO, KG |
003y02bora | Bora | III | 2002 | f | H27 | B. Randrianambinina, S. Rasoloharijaona |
002y02bora | Bora | III | 2002 | m | H4 | B. Randrianambinina, S. Rasoloharijaona |
001y02bora | Bora | III | 2002 | m | H4 | B. Randrianambinina, S. Rasoloharijaona |
Rights and permissions
About this article
Cite this article
Guschanski, K., Olivieri, G., Funk, S.M. et al. MtDNA reveals strong genetic differentiation among geographically isolated populations of the golden brown mouse lemur, Microcebus ravelobensis . Conserv Genet 8, 809–821 (2007). https://doi.org/10.1007/s10592-006-9228-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-006-9228-4