Abstract
Multiple receptors conformation docking (MRCD) and clustering of dock poses allows seamless incorporation of receptor binding conformation of the molecules on wide range of ligands with varied structural scaffold. The accuracy of the approach was tested on a set of 120 cyclic urea molecules having HIV-1 protease inhibitory activity using 12 high resolution X-ray crystal structures and one NMR resolved conformation of HIV-1 protease extracted from protein data bank. A cross validation was performed on 25 non-cyclic urea HIV-1 protease inhibitor having varied structures. The comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) models were generated using 60 molecules in the training set by applying leave one out cross validation method, r 2loo values of 0.598 and 0.674 for CoMFA and CoMSIA respectively and non-cross validated regression coefficient r2 values of 0.983 and 0.985 were obtained for CoMFA and CoMSIA respectively. The predictive ability of these models was determined using a test set of 60 cyclic urea molecules that gave predictive correlation (r 2pred ) of 0.684 and 0.64 respectively for CoMFA and CoMSIA indicating good internal predictive ability. Based on this information 25 non-cyclic urea molecules were taken as a test set to check the external predictive ability of these models. This gave remarkable out come with r 2pred of 0.61 and 0.53 for CoMFA and CoMSIA respectively. The results invariably show that this method is useful for performing 3D QSAR analysis on molecules having different structural motifs.

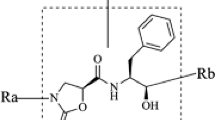
Schematic representation of the multiple receptor conformation docking, clustering and 3D QSAR. Comparative molecular field analysis (CoMFA) and comparative molecular similarity indices (CoMSIA) analysis is an exceptional tool for understanding the structure activity relations of molecules towards their biological activities. Receptor binding conformation of the molecule gives an added advantage to understand ligand receptor interactions required for bioactivity. There are different methods employed for obtaining the receptor based alignment of the molecules, but this method is limited to molecules having common substructure. Here we describe the new approach of multiple receptors conformation docking (MRCD) and clustering of dock poses that allows seamless incorporation of receptor binding conformation of the molecules on wide range of molecules with varied structural scaffold.









Similar content being viewed by others
References
Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zolla-Pazner S, Leibowitch J, Popovic M (1983) Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science 220:865–867
Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vézinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L (1983) Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871
Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RAF, Scolnick EM, Sigal IS (1988) Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA 85:4686–4690
Tomasselli AG, Howe WJ, Sawyer TK, Wlodawer A, Heinrikson RL (1991) The complexities of AIDS: An assessment of the HIV protease as a Therapeutic Target. Chim Oggi 9:6–27
Huff JR (1991) HIV Protease A Novel chemotherapeutic Target for AIDS. J Med Chem 34:2305–2314
Norbeck DW, Kempf DJ (1991) HIV Protease Inhibitors. Annu Rep Med Chem 26:141–160
Debouck C (1992) The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retroviruses 8:153–164
Katz RA, Skalka AM (1994) The Retroviral Enzymes. Annu Rev Biochem 63:133–173
Lam PYS, Jadhav PK, Eyermann CJ, Hodge CN, Ru Y, Bacheler LT, Meek JL, Otto MJ, Rayner MM, Wong YN, Chang CH, Weber PC, Sharpe TR, Jackson DA, Erickson-Viitanen S (1994) Rational design of potent bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science 263:380–384
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146
Kubinyi H, Folkers G, Martin YC (eds) (1998) 3D-QSAR in drug design, vol 2. Kluwer,Dordrecht, the Netherlands
Veerapandian P (ed) (1997) Structure-Based Drug Design. Dekker, New York
Cho SJ, Garsia MLS, Bier J, Tropsha A (1996) Structure-Based Alignment and Comparative Molecular Field Analysis of Acetylcholinesterase Inhibitors. J Med Chem 39:5064–5071
Sippl W, Contreras JM, Parrot I, Rival YM, Wermuth CG (2001) Structure-based 3D QSAR and design of novel acetylcholinesterase inhibitors. J Comput-Aided Mol Des 15:395–410
Sippl W (2000) Receptor-based 3D QSAR analysis of estrogen receptor ligands – merging the accuracy of receptor-based alignments with the computational efficiency of ligand-based methods. J Comput-Aided Mol Des 14:559–572
McGovern DL, Mosier PD, Roth BL, Westkaemper RBK (2010) CoMFA analyses of C-2 position Salvinorin A analogs at the kappa-opioid receptor provides insights into epimer selectivity. J Mol Graph Model 28:612–625
Muddassar M, Pasha FA, Chung HW, Yoo KH, Oh CH, Cho SJ (2008) Receptor guided 3D-QSAR: a yseful approach for designing of IGF-1R inhibitors. J Biomed Biotechnol, vol. 2008, Article ID 837653. doi:10.1155/2008/837653
Jojart B, Marki A (2007) Receptor-based QSAR studies of non-peptide human oxytocin receptor antagonists. J Mol Graph Model 25:711–720
Carlson HA, Masukawa KM, McCammon JA (1999) Method for including the dynamic fluctuations of a protein in computer-aided drug design. J Phys Chem 103:10213–10219
Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, Briggs JM, McCammon JA (2000) Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem 43:2100–2114
Kanth SS, Vijjulatha M (2008) Tetrahydroxy cyclic urea-potent inhibitor for HIV-1 protease wild type and mutant type - a computational design. E-J Chem 5:584–592
Sham HL, Zhao C, Stewart KD, Betebenner DA, Lin S, Park CH, Kong XP, Rosenbrook W Jr, Herrin T, Madigan D, Vasavanonda S, Lyons N, Molla A, Saldivar A, Marsh KC, McDonald E, Wideburg NE, Denissen JF, Robins T, Kempf DJ, Plattner JJ, Norbeck DW (1996) A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease. J Med Chem 39:392–397
Ala PJ, Huston EE, Klabe RM, Jadhav PK, Lam PY, Chang CH (1998) Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities. Biochemistry 37:15042–15049
Backbro K, Lowgren S, Osterlund K, Atepo J, Unge T, Hulten J, Bonham NM, Schaal W, Karlen A, Hallberg A (1997) Unexpected binding mode of a cyclic sulfamide HIV-1 protease inhibitor. J Med Chem 40:898–902
Huang PP, Randolph JT, Klein LL, Vasavanonda S, Dekhtyar T, Stoll VS, Kempf DJ (2004) Synthesis and antiviral activity of P1' arylsulfonamide azacyclic urea HIV protease inhibitors. Bioorg Med Chem Lett 14:4075–4078
Jadhav PK, Ala P, Woerner FJ, Chang CH, Garber SS, Anton ED, Bacheler LT (1997) Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV. J Med Chem 40:181–191
Lam PY, Ru Y, Jadhav PK, Aldrich PE, DeLucca GV, Eyermann CJ, Chang CH, Emmett G, Holler ER, Daneker WF, Li L, Confalone PN, McHugh RJ, Han Q, Li R, Markwalder JA, Seitz SP, Sharpe TR, Bacheler LT, Rayner MM, Klabe RM, Shum L, Winslow DL, Kornhauser DM, Hodge CN (1996) Cyclic HIV protease inhibitors: synthesis, conformational analysis, P2/P2' structure-activity relationship, and molecular recognition of cyclic ureas. J Med Chem 39:3514–3525
Jadhav PK, Woerner FJ, Lam PY, Hodge CN, Eyermann CJ, Man HW, Daneker WF, Bacheler LT, Rayner MM, Meek JL, Erickson-Viitanen S, Jackson DA, Calabrese JC, Schadt M, Chang CH (1998) Nonpeptide cyclic cyanoguanidines as HIV-1 protease inhibitors: synthesis, structure-activity relationships, and X-ray crystal structure studies. J Med Chem 41:1446–1455
Hodge CN, Aldrich PE, Bacheler LT, Chang CH, Eyermann CJ, Garber S, Grubb M, Jackson DA, Jadhav PK, Korant B, Lam PY, Maurin MB, Meek JL, Otto MJ, Rayner MM, Reid C, Sharpe TR, Shum L, Winslow DL, Erickson-Viitanen S (1996) Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450. Chem Biol 3:301–314
Schaal W, Karlsson A, Ahlsen G, Lindberg J, Andersson HO, Danielson UH, Classon B, Unge T, Samuelsson B, Hulten J, Hallberg A, Karlen A (2001) Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors. J Med Chem 44:155–169
Yamazaki T, Hinck AP, Wang YX, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Chang CH, Domaille PJ, Lam PY (1996) Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy. Protein Sci 5:495–506
Schrödinger LLC (2005) Glide, Version 4.0. New York, NY
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw ED, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J Med Chem 47:1739–1749
Nugiel DA, Jacobs K, Worley T, Patel M, Kaltenbach RF III, Meyer DT, Jadhav PK, de Lucca GV, Smyser TE, Klabe RM, Bacheler LT, Rayner MM, Seitz SP (1996) Preparation and structure-activity relationship of novel P1/P1‘-substituted cyclic urea-based human immunodeficiency virus type-1 protease inhibitors. J Med Chem 39:2156–2169
Patel M, Kaltenbach RF III, Nugie DA, Mchugh RJ Jr, Jadhav PK, Bacheler LT, Cordova BC, Klabe RM, Erickson-Viitanen S, Garber SS, Ried C, Sitz SP (1998) The synthesis of symmetircal and unsymmetrical P1/P1’ Cyclic ureas as HIV protease inhibitors. Bioorg Med Chem Lett 8:1077–1082
Patel M, Bacheler LT, Rayner MM, Cordova BC, Klabe RM, Erickson Viitanen S, Sitz SP (1998) The synthesis and evaluation of cyclic ureas as HIV protease inhibitors: modification of the P1/P1’ groups. Bioorg Med Chem Lett 8:823–828
Rodgers JD, Johnson BL, Wang H, Erickson-Viitanen S, Klabe RM, Bacheler LT, Cordova BC, Chang CH (1998) Potent cyclic urea HIV protease inhibitors with 3-aminoindazole P2/P2’groups. Bioorg Med Chem Lett 8:715–720
Ax A, Schaal W, Vrang L, Samuelsson B, Hallberg A, Karlén A (2005) Cyclic sulfamide HIV-1 protease inhibitors, with sidechains spanning from P2/P2′ to P1/P1′. Bioorg Med Chem 13:755–764
Hultén J, Andersson HO, Schaal W, Danielson HU, Classon B, Kvarnström I, Karlén A, Unge T, Samuelsson B, Hallberg A (1999) Inhibitors of the C2-symmetric HIV-1 protease: nonsymmetric binding of a symmetric cyclic sulfamide with ketoxime groups in the P2/P2‘ side chains. J Med Chem 42:4054–4061
Sybyl version 6.9 (1999) Tripos Associates, St. Louis (MO)
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity – a rapid access to atomic charges. Tetrahedron 36:3219–3228
Wheelock CE, Nakagawa Y, Harada T, Oikawa N, Akamatsu M, Smagghe G, Stefanou D, Iatrou K, Swevers L (2006) High-throughput screening of ecdysone agonists using a reporter gene assay followed by 3-D QSAR analysis of the molting hormonal activity. Bioorg Med Chem 14:1143–1159
Acknowledgments
We gratefully acknowledge support for this research from Department of Science and Technology, New Delhi, India, University Grants Commission, New Delhi, India and Department of chemistry, Nizam College, Hyderabad, India. We are greatly thankful to Dr. G. N. Shastry, Scientist, Indian Institute of Chemical Technology for Sybyl 6.9 software and his useful suggestions. We also acknowledge Schrödinger Inc. for GLIDE software.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
1.pdb
(TXT 5 kb)
2.pdb
(TXT 6 kb)
3.pdb
(TXT 6 kb)
4.pdb
(TXT 7 kb)
5.pdb
(TXT 8 kb)
6.pdb
(TXT 8 kb)
7.pdb
(TXT 7 kb)
8.pdb
(TXT 7 kb)
9.pdb
(TXT 7 kb)
10.pdb
(TXT 8 kb)
11.pdb
(TXT 8 kb)
12.pdb
(TXT 9 kb)
13.pdb
(TXT 6 kb)
14.pdb
(TXT 7 kb)
15.pdb
(TXT 6 kb)
16.pdb
(TXT 7 kb)
17.pdb
(TXT 7 kb)
18.pdb
(TXT 7 kb)
19.pdb
(TXT 8 kb)
20.pdb
(TXT 7 kb)
21.pdb
(TXT 7 kb)
22.pdb
(TXT 7 kb)
23.pdb
(TXT 7 kb)
24.pdb
(TXT 9 kb)
25.pdb
(TXT 7 kb)
26.pdb
(TXT 7 kb)
27.pdb
(TXT 7 kb)
28.pdb
(TXT 7 kb)
29.pdb
(TXT 7 kb)
30.pdb
(TXT 7 kb)
31.pdb
(TXT 7 kb)
32.pdb
(TXT 8 kb)
33.pdb
(TXT 8 kb)
34.pdb
(TXT 8 kb)
35.pdb
(TXT 8 kb)
36.pdb
(TXT 8 kb)
37.pdb
(TXT 8 kb)
38.pdb
(TXT 8 kb)
39.pdb
(TXT 8 kb)
40.pdb
(TXT 9 kb)
41.pdb
(TXT 10 kb)
42.pdb
(TXT 10 kb)
43.pdb
(TXT 10 kb)
44.pdb
(TXT 11 kb)
45.pdb
(TXT 10 kb)
46.pdb
(TXT 10 kb)
47.pdb
(TXT 9 kb)
48.pdb
(TXT 11 kb)
49.pdb
(TXT 10 kb)
50.pdb
(TXT 10 kb)
51.pdb
(TXT 10 kb)
52.pdb
(TXT 11 kb)
53.pdb
(TXT 11 kb)
54.pdb
(TXT 10 kb)
55.pdb
(TXT 10 kb)
56.pdb
(TXT 10 kb)
57.pdb
(TXT 11 kb)
58.pdb
(TXT 10 kb)
59.pdb
(TXT 10 kb)
60.pdb
(TXT 10 kb)
61.pdb
(TXT 10 kb)
62.pdb
(TXT 10 kb)
63.pdb
(TXT 11 kb)
64.pdb
(TXT 10 kb)
65.pdb
(TXT 9 kb)
66.pdb
(TXT 9 kb)
67.pdb
(TXT 10 kb)
68.pdb
(TXT 10 kb)
69.pdb
(TXT 11 kb)
70.pdb
(TXT 10 kb)
71.pdb
(TXT 9 kb)
72.pdb
(TXT 8 kb)
73.pdb
(TXT 9 kb)
74.pdb
(TXT 10 kb)
75.pdb
(TXT 5 kb)
76.pdb
(TXT 9 kb)
77.pdb
(TXT 8 kb)
78.pdb
(TXT 7 kb)
79.pdb
(TXT 7 kb)
80.pdb
(TXT 8 kb)
81.pdb
(TXT 8 kb)
82.pdb
(TXT 9 kb)
83.pdb
(TXT 8 kb)
84.pdb
(TXT 9 kb)
85.pdb
(TXT 8 kb)
86.pdb
(TXT 8 kb)
87.pdb
(TXT 8 kb)
88.pdb
(TXT 8 kb)
89.pdb
(TXT 8 kb)
90.pdb
(TXT 8 kb)
91.pdb
(TXT 9 kb)
92.pdb
(TXT 8 kb)
93.pdb
(TXT 7 kb)
94.pdb
(TXT 8 kb)
95.pdb
(TXT 9 kb)
96.pdb
(TXT 10 kb)
97.pdb
(TXT 6 kb)
98.pdb
(TXT 6 kb)
99.pdb
(TXT 6 kb)
100.pdb
(TXT 7 kb)
101.pdb
(TXT 7 kb)
102.pdb
(TXT 7 kb)
103.pdb
(TXT 7 kb)
104.pdb
(TXT 8 kb)
105.pdb
(TXT 6 kb)
106.pdb
(TXT 7 kb)
107.pdb
(TXT 9 kb)
108.pdb
(TXT 8 kb)
109.pdb
(TXT 8 kb)
110.pdb
(TXT 8 kb)
111.pdb
(TXT 7 kb)
112.pdb
(TXT 8 kb)
113.pdb
(TXT 7 kb)
114.pdb
(TXT 9 kb)
115.pdb
(TXT 10 kb)
116.pdb
(TXT 9 kb)
117.pdb
(TXT 10 kb)
118.pdb
(TXT 12 kb)
119.pdb
(TXT 9 kb)
120.pdb
(TXT 10 kb)
ncu1.pdb
(TXT 4 kb)
ncu2.pdb
(TXT 7 kb)
ncu3.pdb
(TXT 4 kb)
ncu4.pdb
(TXT 4 kb)
ncu5.pdb
(TXT 7 kb)
ncu6.pdb
(TXT 6 kb)
ncu7.pdb
(TXT 7 kb)
ncu8.pdb
(TXT 7 kb)
ncu9.pdb
(TXT 5 kb)
ncu10.pdb
(TXT 7 kb)
ncu11.pdb
(TXT 8 kb)
ncu12.pdb
(TXT 8 kb)
ncu13.pdb
(TXT 8 kb)
ncu14.pdb
(TXT 7 kb)
ncu15.pdb
(TXT 8 kb)
ncu16.pdb
(TXT 8 kb)
ncu17.pdb
(TXT 8 kb)
ncu18.pdb
(TXT 7 kb)
ncu19.pdb
(TXT 8 kb)
ncu20.pdb
(TXT 8 kb)
ncu21.pdb
(TXT 8 kb)
ncu22.pdb
(TXT 8 kb)
ncu23.pdb
(TXT 8 kb)
ncu24.pdb
(TXT 8 kb)
ncu25.pdb
(TXT 5 kb)
Rights and permissions
About this article
Cite this article
Sivan, S.K., Manga, V. Multiple receptor conformation docking and dock pose clustering as tool for CoMFA and CoMSIA analysis – a case study on HIV-1 protease inhibitors. J Mol Model 18, 569–582 (2012). https://doi.org/10.1007/s00894-011-1048-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1048-x