Abstract
Human ethnic groups are frequently comprised of two or more founder populations. One of these founding populations is often available for contemporary sampling. We describe a method for reconstructing the composition of a missing founder population using the highly informative haplotypes comprising the HLA system. An application of the method is demonstrated using bone marrow registry samples of African Americans. We use contemporary samples of African Americans and European Americans to derive haplotypes of the West African founder populations. This approach may also be useful for reconstructing ancestral haplotypes for regions elsewhere in the genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During human history, the process of spreading over first Africa and then the other continents divided humans into more or less discrete populations defined by distinct cultural practices. Periods of separation, selection and population isolation laid conditions for varying degrees of genetic differentiation (e.g. Johansson and Gyllensten 2008). The re-contact and admixture of these more or less discrete groups has resulted in new distinct populations comprised of two or more founding groups. Such admixed groups often constitute well-recognized ethnic categories in countries throughout the world.
Historically, recent admixture from populations of differing continental origins defines such groups in the USA and Brazil, for example. Yet the admixture of groups from intra-continental sources has also been a familiar aspect of human population history. For example, the largest human population group, the Han Chinese, have been shown to consist of distinct subpopulations reflecting diverse origins, partial isolation and subsequent admixture (Hu et al. 2007; Chen et al. 2008). Even within national boundaries, European populations are also the product of waves of invasions and subsequent admixture reflected in their current genetic composition. As we demonstrate here, it is straightforward and unambiguous to reconstruct greatly diverged parental populations.
The HLA system, including the loci of the human major histocompatibility complex at 6p21, comprises the most polymorphic system in the human genome. It evolves rapidly and as such constitutes an excellent marker system for identifying and following the parental population contributions to contemporary admixed groups. Given the HLA-typed samples of an admixed population and one of its founder populations, a method to identify the HLA haplotypes of the missing founder population is described and applied.
Methods
Model for division of samples
If two founder populations PA and PB admix to produce a new admixed population, PN (Fig. 1), then the frequencies of each HLA haplotype i will be related:
where M is the proportion of PA in the new population PN. Given that PA and PN can be sampled and typed, then the frequency of each haplotype PBi in population PB can be expressed:
In this formulation, the admixture rate M is estimated from outside sources. The preferred sources for admixture estimates are genomewide studies with pure parental population controls (e.g. Price et al. 2007; Risch 2006). The geographical scale of the estimate should be considered. For example, African American composition has been shown to vary across the United States, with African Americans from the US South having a higher fraction of African background genetics. Large-scale estimates can be achieved by averaging a set of small-scale studies in different locales to get a nationwide average, but care should be taken when using estimates from smaller population and sample locations.
While the vast majority of haplotypes are private to different continental populations, some haplotypes are found in both populations, but at differing frequencies. Our method handles haplotypes both shared and private.
Sampling variation may result in negative frequencies when the product of the admixture proportion M and the frequency of the haplotype in population PA exceeds the frequency of the haplotype in the admixed population PN. In these cases, the haplotype is estimated not to exist in PB, so the frequency of the haplotype in PB is set to zero. After the initial determination of all haplotype frequencies, PBi, all haplotypes are normalized to sum to one.
Selecting pure ancestral population samples
Once the ancestral population frequencies have been calculated, it is possible to split the list of genotypes from the admixed population sample into separate categories for individuals with pure ancestry and those with mixed ancestry. This has applications in disease association studies for generating sets of case/control populations with the same ancestral background. We have used a Bayesian approach using the ancestral haplotype frequencies to assign an ancestral label to the haplotype of each individual. The weighting parameters are adjusted until the desired admixture proportion is reached. We consider the admixture proportion at the haplotype level to count individuals with mixed ancestry towards the total level of admixture (Eq. 2).
Estimating admixture from HLA data
We can calculate admixture proportions using HLA data when no estimate is currently available from other sources. With all source populations characterized, one can calculate the relative contribution in an admixed population where admixture proportion is unknown. The solution is to solve for the admixture proportion where the difference between the linear combination of source populations and the admixed population is minimized (Eq. 3).
In admixed populations with more than two founder populations, such as Caribbean Hispanic populations that have a mixture of African, Native American and European ancestry, the same method can be applied to calculate the frequencies of a single missing founder population when all the other founder populations have been characterized and admixture estimates provided for each component.
Population data
The National Marrow Donor Program in Minneapolis, MN maintains a donor registry including individuals of African Americans and of European Americans. A total of 1,000 individuals from each registry group were randomly selected. We utilized National Marrow Donor Program data recently reported in the literature (Maiers et al. 2007).
HLA typing and haplotype inference
HLA typing was performed at the antigen or two-digit level of resolution at the loci HLA A, HLA B and HLA DRB1 using DNA methods. Three locus haplotypes, for example A*32-B*42-DRB1*03, are abbreviated 32:42:03.
For haplotype inference, we used standard methods adjusted to accommodate the large haplotype diversity present in the HLA system. We applied the expectation-maximization (EM) algorithm to infer three locus HLA haplotypes from genotypes. Estimation of frequencies of rare haplotypes in founder populations is highly prone to error. There are several sources of possible error. Inadequate sampling of populations results in frequencies that have a wide error bounds due to statistical variation in sampling a small proportion of the overall population. Estimation error is an artifact of the EM algorithm where rare haplotypes in the sampled populations are difficult to ascertain due to lack of information. Admixture estimation error affects the frequencies of the missing founder population calculation based on the accuracy of the admixture proportion into the admixed population. Some haplotypes may have been created by recombination or mutation after the merging of the two founder populations. This method assigns these haplotypes to the missing founder population.
Application
The method of derivation of founder population haplotypes can be demonstrated with HLA-typed samples of European Americans and African Americans. African Americans are derived from West Africans and Europeans in the proportions of approximately 80:20 (e.g. Zhu et al. 2005). This example is especially informative for these purposes because of the great (intercontinental) divergence in HLA haplotypes between the peoples of Africa and Europe (Mack and Erlich 2006). Infrequent but genuine haplotype similarities or the possibility of low levels of African Admixture in the European American sample (Shriver et al. 2003) will not detract from the utility of this example because of the substantial differences between the two founding populations. In order to estimate the HLA haplotype frequencies of the West African founder populations, we took samples of 1,000 individuals (2,000 haplotypes) typed at the “antigen level” (2-digit) for African American and European American donor samples from the National Marrow Donor Program registries (Maiers et al. 2007).
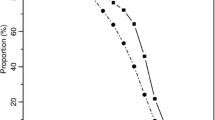
Estimated three locus haplotypes were sorted by frequency and the ten most common haplotypes displayed by order of each of the two founder populations along with the second founder population and the admixed (African American) population sample (Fig. 2a, b). The most common European American haplotype, A*01-B*08-DRB1*03, present at a frequency of 0.067 in the European American sample, was present in the African Americans at a frequency of only 0.007 or 10.4% of that seen in European Americans. It was entirely absent in the derived West Africans. For the top ten haplotypes the African American frequencies were present near 20% (mean = 20.7%) of the frequencies observed in the European Americans (Fig. 2a). The derived West African A-B-DRB1 haplotypes were in fact not observed in the European Americans at four of the ten possible instances, i.e. detectable at a frequency of ≥0.001.
a Frequencies of the 10 most common European HLA haplotypes (EUR) plotted against African American (NAFA) and derived West African (AFR) haplotype frequencies, b Frequencies of the 10 most common derived West African HLA haplotypes plotted against African American and European American haplotype frequencies
The most frequent ten West African HLA haplotypes similarly arranged and compared with the frequencies of the African American and European American haplotypes are shown in Fig. 2b. The most common West African haplotype is A*30-B*42-DRB1*03, present at a frequency of 0.021. African American frequencies averaged 92% that of the estimated West African frequencies. The two European American haplotypes observed in the West African sample were quite rare and may be due to haplotype estimation errors with frequencies of only 0.00054 and 0.00026 for haplotypes 23:15:11 and 74:15:13, respectively.
Although a fuller description of founder populations estimated from African Americans and other groups will be presented separately, some points are worth making at this time. This example, comparing differences in HLA frequencies between two continental regions, suggests that there may be complete population differentiation in HLA types at the continental level, with little or no sharing of haplotypes. Further underlining this point, the two-digit antigen level of HLA typing resolution presented here often contains a great deal of additional allelic variation, which can make a sizeable contribution to haplotypic variation. For example, the common alleles seen in Europeans, B*44 and DRB1*15, each consist of dozens of subtypes. An additional source of further HLA haplotype variation is present in the other histocompatibility loci also present in the HLA complex. We suggest that samples typed at high resolution and at addition HLA loci would further reduce instances of haplotype overlap between European and African source HLA haplotypes.
Discussion
Historically admixed populations have gained attention in recent years because of their potential for admixture mapping of disease genes (Smith and O-Brien 2005; Patterson et al. 2004; Wang et al. 2008; Xu et al. 2008). Our goal in this contribution is to demonstrate a method to re-create parental populations of an admixed group, when one of the parental populations is available, especially pertinent to HLA information. The HLA region of humans is composed of the highly polymorphic major histocompatibility loci distributed over a region of 3–4 Mb. The high diversity of this region is much greater than the sum of the allelic variation from each of the 8–10 histocompatibility loci.
Population samples of HLA frequencies derived by this method can be of value in several respects. First, one or more of the founder populations of a contemporary group will often be unavailable or impossible to sample, making the reconstructed samples of unique value. In addition, a population’s HLA composition is an essential starting place for determining the sampling requirements for an ethnically specific bone marrow or stem cell registry, and in understanding the practical side of population differentiation for patient–donor matching. It is possible to stratify admixed groups based on inference of their HLA haplotypes coming from two ancestral sources or a single population. Patients with mixed ancestral HLA will be among the least likely to find a match because population samples with similar ancestral mixtures may be difficult to obtain.
This work describes a method of reconstructing the haplotype frequencies of a founder population. For this purpose, we use only a relatively small sample of available population data (1,000 European Americans and 1,000 African Americans) and limit the description of haplotypes from the derived population. A more complete and thorough study founder HLA haplotypes from African American and other admixed populations will be reported separately, and will address relative subtleties of the method such as the adequacy and purity of an available founder population (e.g. European Americans) and present more substantial lists of derived haplotypes. Another issue to be addressed at that time is the apparent sharing of haplotypes from populations of intercontinental origins.
The evolution and modification of haplotypes of the HLA complex have been studied over many years, yet haplotype blocks present throughout the genome also evolve through the same variety of genetic mechanisms as seen in the HLA system. Our method applies to not just the HLA system, but to other haplotype frequencies in the genome. The HLA system is particularly remarkable for the availability of quality data and the population privacy of its haplotypes. Datasets are often available using other genetic marker systems, raising the possibility for this same type of analysis on SNPs or microsatellites. In fact, forays have already been made into analyzing admixed populations with a variety of genetic systems (Bertorelle and Excoffier 1998; Mountain et al. 2002; Choisy et al. 2004; Pfaff et al. 2004; Price et al. 2007). It appears that the HLA system may one end of the spectrum of population haplotype divergence in humans.
References
Bertorelle G, Excoffier L (1998) Inferring admixture proportions from molecular data. Mol Biol Evol 15:1298–1311
Chen L, Qin S, Xie J, Tang J, Yang L, Shen W, Zhao X, Du J, He G, Feng F, He L, Qinghe X (2008) Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographic areas of mainland China. Pharmacogenomics 9:601–602
Choisy M, Franck P, Cornuet J-M (2004) Estimating admixture proportions with microsatellites: comparison of methods based on simulated data. Mol Ecol 13:955–968
Hu S-P, Luan J-A, Li B, Chen J-X, Cai K-L, Huang L-Q, Xu X-Y (2007) Genetic link between Chaoshan and other Chinese Han populations: Evidence from the HLA-A and HLA-B allele frequency distribution. Am J Phys Anthropol 143:140–150
Johansson A, Gyllensten U (2008) Identification of local selective sweeps in human populations since the exodus from Africa. Hereditas 145:126–137
Mack S, Erlich H (2006) Population relationships as inferred from classical HLA genes. 13th International Histocompatibility Workshop Anthropology/Human Genetic Diversity Joint Report in Immunobiology of the Human MHC. Proceedings of the 13th international histocompatibility workshop and congress. Fred Hutchinson Cancer Research Center Seattle WA, USA
Maiers M, Gragert L, Klitz W (2007) High resolution HLA alleles and haplotypes in the US population. Hum Immunol 68:779–788
Mountain JL, Knight A, Jobin M, Gignoux C, Miller A, Lin AA, Underhill PA (2002) SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res 12:1766–1772
Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D (2004) Methods for high-density admixture mapping of disease genes. Am J Hum Genet 74:979–1000
Pfaff CL, Barnholtz-Sloan J, Wagner JK, Long JC (2004) Information on ancestry from genetic markers. Genet Epidemiol 26:3005–3315
Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D (2007) A genomewide admixture map for latino populations. Am J Hum Genet 80:1024–1036
Risch N (2006) Dissecting racial and ethnic differences. N Engl J Med 354:408–411
Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA (2003) Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 112:387–399
Smith MW, O-Brien SJ (2005) Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet 6:623–632
Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Polleti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer N, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro EI, Bailliet G, Bianchi NL, Llop E, Rothammer F, Excoffier L, Ruiz-Linares A (2008) Geographic patterns of genome admixture in Latin American mestizos. PLoS Genet 4(3):e1000037
Xu S, Huang W, Qian J, Jin L (2008) Analysis of genomic admixture in Uhghur and its implication in mapping strategy. Am J Hum Genet 82:883–894
Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A (2005) Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 37:177–181
Acknowledgments
Supported in part by Office of Naval Research Grant N00014-08-1-0058.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Klitz, W., Maiers, M. & Gragert, L. Re-creation of the genetic composition of a founder population. Hum Genet 124, 417–421 (2008). https://doi.org/10.1007/s00439-008-0565-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-008-0565-x