Abstract
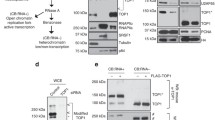
Conjugation of SUMO to target proteins is an essential eukaryotic regulatory pathway. Multiple potential SUMO substrates were identified among nuclear and chromatin proteins by proteomic approaches. However, the functional roles of SUMO-modified pools of individual proteins remain largely obscure, as only a small fraction of a given target is sumoylated and therefore is experimentally inaccessible. To overcome this technical difficulty in case of Topoisomerase II, we employed constitutive SUMO modification, enabling tracking of modified Top2p, not only biochemically but also cytologically and genetically. Topoisomerase II fused to a critical number of SUMO repeats is concentrated at the specific intranuclear domain, the nucleolus, when more than four SUMO moieties are added, indicating that fused SUMO repeats are biologically active. Further analysis has established that poly-sumoylation of Top2p is required for the stable maintenance of the nucleolar organizer, linking SUMO-mediated targeting to functional maintenance of ribosomal RNA gene cluster.





Similar content being viewed by others
References
Bachant J, Alcasabas A et al (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 9(6):1169–1182
Brill SJ, DiNardo S et al (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326(6111):414–416
Christman MF, Dietrich FS et al (1988) Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55(3):413–425
D’Amours D, Stegmeier F et al (2004) Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117(4):455–469
Denison C, Rudner AD et al (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4(3):246–254
Dobreva G, Dambacher J et al (2003) SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev 17(24):3048–3061
Hannich JT, Lewis A et al (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280(6):4102–4110
Hecker CM, Rabiller M et al (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281(23):16117–16127
Huang TT, Wuerzberger-Davis SM et al (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115(5):565–576
Huh WK, Falvo JV et al (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686–691
Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73:355–382
Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272(43):26799–26802
Johnson ES, Blobel G (1999) Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol 147(5):981–994
Kerscher O (2007) SUMO junction—what’s your function? New insights through SUMO-interacting motifs. EMBO Rep 8(6):550–555
Kim RA, Wang JC (1989) A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57(6):975–985
Li SJ, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20(7):2367–2377
Lin DY, Huang YS et al (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24(3):341–354
Mahajan R, Delphin C et al (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88(1):97–107
Matunis MJ, Coutavas E et al (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135(6 Pt 1):1457–1470
Mo YY, Yu Y et al (2002) Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem 277(4):2958–2964
Montpetit B, Hazbun TR et al (2006) Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol 174(5):653–663
Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32(6):286–295
Panse VG, Hardeland U et al (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279(40):41346–41351
Reindle A, Belichenko I et al (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci 119(Pt 22):4749–4757
Ross S, Best JL et al (2002) SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell 10(4):831–842
Saeki Y, Isono E et al (2004) Intracellularly inducible, ubiquitin hydrolase-insensitive tandem ubiquitins inhibit the 26S proteasome activity and cell division. Genes Genet Syst 79(2):77–86
Saitoh H, Sparrow DB et al (1998) Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol 8(2):121–124
Sakaguchi A, Akashi T et al (2001) A distinct subnuclear localization of mammalian DNA topoisomerase IIbeta in yeast. Biochem Biophys Res Commun 283(4):876–882
Sasaki T, Toh EA et al (2000) Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol Cell Biol 20(21):7971–7979
Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100(23):13225–13230
Shilatifard A (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75:243–269
Strunnikov AV, Aravind L et al (2001) Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158(1):95–107
Sun H, Leverson JD et al (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. Embo J 26:4102–4112
Takahashi Y, Toh EA et al (2003) Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J Biochem (Tokyo) 133(4):415–422
Takahashi Y, Yong-Gonzalez V et al (2006) SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics 172(2):783–794
Torres-Rosell J, Sunjevaric I et al (2007) The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9(8):923–931
Uzunova K, Gottsche K et al (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282:34167–34175
Varshavsky A (2005) Ubiquitin fusion technique and related methods. Methods Enzymol 399:777–799
Wang BD, Butylin P et al (2006) Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle 5(19):2260–2267
Wang BD, Eyre D et al (2005) Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol 25(16):7216–7225
Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3(6):430–440
Wohlschlegel JA, Johnson ES et al (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem 279(44):45662–45668
Wykoff DD, O’Shea EK (2005) Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics 4(1):73–83
Xue Y, Zhou F et al (2006) SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res 34:W254–W257 (Web Server issue)
Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102(13):4777–4782
Zhou W, Ryan JJ et al (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279(31):32262–32268
Acknowledgments
Authors thank Y. Kikuchi for plasmids and strains, V. Yong-Gonzalez, P. Butylin, S. Dulev, A. Samoshkin, and X. Strunnikova for fruitful discussion, and S. Malhotra and Y. Ejamo for technical assistance. This work was supported by the NICHD Intramural Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg
Rights and permissions
About this article
Cite this article
Takahashi, Y., Strunnikov, A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma 117, 189–198 (2008). https://doi.org/10.1007/s00412-007-0137-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-007-0137-1