Abstract
A well-documented pharmacometric (PMx) analysis dataset specification ensures consistency in derivations of the variables, naming conventions, traceability to the source data, and reproducibility of the analysis dataset. Lack of standards in creating the dataset specification can lead to poor quality analysis datasets, negatively impacting the quality of the PMx analysis. Standardization of the dataset specification within an individual organization helps address some of these inconsistencies. The recent introduction of the Clinical Data Interchange Standards Consortium (CDISC) Analysis Data Model (ADaM) Population Pharmacokinetic (popPK) Implementation Guide (IG) further promotes industry-wide standards by providing guidelines for the basic data structure of popPK analysis datasets. However, manual implementation of the standards can be labor intensive and error-prone. Hence, there is still a need to automate the implementation of these standards. In this paper, we present PmWebSpec, an easily deployable web-based application to facilitate the creation and management of CDISC-compliant PMx analysis dataset specifications. We describe the application of this tool through examples and highlight its key features including pre-populated dataset specifications, built-in checks to enforce standards, and generation of an electronic Common Technical Document (eCTD)-compliant data definition file. The application increases efficiency, quality and semi-automates PMx analysis dataset, and specification creation and has been well accepted by pharmacometricians and programmers internally. The success of this application suggests its potential for broader usage across the PMx community.
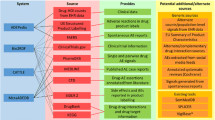
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High-quality dataset specifications are the foundation of a robust pharmacometric (PMx) analysis dataset. It not only ensures the inclusion of the correct variables in the PMx analysis, but also plays a crucial role in enabling traceability and reproducibility, enhancing the reliability and confidence in the analysis results (1, 2).
Currently, dataset specifications are being created manually by pharmacometricians and programmers which can lead to inconsistencies across analyses. PMx analyses often require pooling data from multiple studies and can be very challenging and time-consuming, especially when individual datasets were created using different standards. As many requirements and imputation rules can be shared across projects (3), there is a need to enforce uniform standards in dataset specifications. Standardization of dataset specifications improves dataset quality, minimizes the effort needed for review and validation, facilitates automation of dataset creation, and streamlines subsequent analyses.
The need for a standardized PMx analysis dataset specification is underscored by the recent introduction of the Clinical Data Interchange Standards Consortium (CDISC) Analysis Data Model (ADaM) Population Pharmacokinetic (popPK) Implementation Guide (IG) for popPK analysis (4). A dataset specification should be created with the objective of being “analysis-ready”, containing the variables needed for the intended use of popPK analysis: subject identifier variables, event variables, time variables, treatment variables, and covariates. The IG provides general naming conventions for variables and defines if a variable is required, conditionally required, or permissible along with other variable attributes. While standards exist, there is still a need for tools that can automatically enforce these standards and best practices.
We present PmWebSpec (5), a novel web-based application that automates the creation of analysis dataset specifications and addresses the issues that come with lack of following standards. We demonstrate the features of the application by providing an example of the creation and management of a CDISC-compliant popPK dataset specification, while highlighting the built-in features that enforce quality checks on variable names and attributes. This tutorial describes additional features of the application that facilitate various aspects of the PMx analysis dataset and specification development.
PmWebSpec Overview
Dataset Specification
A high-quality dataset specification should include comprehensive instructions on how to construct a dataset. Although the specific content of the dataset specification may vary across companies and functions, the instructions should at least consist of the dataset structure, a list of required variables and their attributes, identification of source data, derivations, and imputation rules. Additional information such as the locations of source data and codes is not mandatory but can be beneficial for programmers in tracking source data snapshots and managing projects to ensure traceability. To encompass all aspects of the dataset requirements and project background, we have designed five sections in the dataset specification: Specification Information, General Information, Dataset Structure, Derivations, and Confirmations. Furthermore, we have implemented built-in templates and checks to ensure the dataset specification’s quality and integrity.
PmWebSpec templates are pre-populated dataset specifications that include commonly used variables, flags, derivations, and imputations for specific analyses. For example, a popPK template is the initial step to develop a popPK dataset specification. To enforce the CDISC ADaM popPK IG (4), we have created the PPK-CDISC template. All variables in the template are predefined and conform to the IG, which ensures that the minimal requirements of a popPK dataset are met. Table I lists some common CDISC ADaM variables for popPK analysis. The template includes standard flags for record identification, such as day 1 pre-dose samples, post-first dose samples that fall below the limit of quantification, and records with data issues and imputations. Similarly, an Exposure–Response (E-R) template can be used to develop an E-R dataset specification. As a best practice, the E-R template follows the same naming conventions for common variables across popPK and E-R. These templates ensure consistency in dataset specifications across projects and studies, maintain compliance with required standards, and reduce back-and-forth communications between pharmacometricians and programmers. Users have the flexibility to modify existing templates or create their own to accommodate any type of dataset.
Specification Information
The Specification Information section is designed to collect metadata such as compound name and indication. The dataset type, user’s full name, and creation date are automatically populated based on the template type, the logged-in user, and the current date. This metadata is used to generate a specification ID, which serves as a unique identifier within PmWebSpec. The specification ID can be used to search for a dataset specification.
General Information
The General Information section is comprised of text fields where users can enter essential project information, including a concise project description, the purpose of the project, key personnel, source data locations, paths for program development and quality control (QC), dataset attributes, and dataset inclusion criteria (Fig. 1).
The source data location documents the provenance of the data used to construct the dataset. Dataset attributes encompass the dataset name, label, sorting variables, and single/multiple records per subject. This application ensures that the dataset name and label adhere to the electronic Common Technical Document (eCTD) guidelines (6). Dataset inclusion criteria, although often overlooked, are crucial for dataset construction as data pooling is typically required in PMx analysis. It is of utmost importance to explicitly list all studies and cohorts that should be included in the dataset. The inclusion criteria can be utilized to filter and find specifications that include specific studies. Users will be alerted by built-in checks if they omit any mandatory fields.
Dataset Structure
The Dataset Structure section details variable attributes: variable name, label, type, unit, rounding, missing values, notes, and source. The Dataset Structure consists of two tables, one for required variables (Fig. 2A) and another for optional variables (Fig. 2B). The required variable table is automatically populated with the variables that are required in the dataset, based on the template selected.
The optional variable table contains common variables that are not essential for analysis. The attributes of the variables included in this table are predefined and adhere to CDISC standards. These variables can be added to the required variable table by ticking the checkbox next to the variable. To ensure self-documentation within the dataset, specific pairs of character and numeric variables, such as ARACE and ARACEN in the optional table, will both be added to the required variable table.
If variables do not exist in either the required or optional variable table, additional variables can be added using the “Add new variable” button. The attributes of these variables are completely user-defined but the name and label must still conform to eCTD guidelines, which are verified by PmWebSpec. The “Search Variable” button can be used to find variables within other specifications, aiding in creation of user-defined variables (Fig. 2A).
Users have the ability to modify the order of the variables in the required table and delete any optional or user-defined variables. However, users can not modify or delete required variables. The variable attributes and variable order presented in the dataset specification should match the dataset.
Derivations
The Derivations section documents the formulas, derivations, algorithms, and imputations used in the dataset construction. To maintain accuracy and transparency, it is essential to specify the formula used when deriving variables. The CDISC ADaM popPK IG recommends this information to be included in the submission documentation (4). This application allows users to save default formulas and automatically populate them in the derivation table in the dataset specification (as shown in Fig. 3A). Utilizing the default formulas ensures consistency in derivations, which simplifies the process of pooling multiple studies. Additionally, users can add their own formulas to the derivation table.
In PMx analysis datasets, it is common to impute missing values, such as dose date and/or time, resulting from incomplete source data. To identify records with imputed values, it is necessary to include flags in the dataset and thoroughly document the imputation algorithms in the dataset specification. Additional exclusion or information flags can be incorporated into the dataset specification and dataset to identify data points with issues or that need to be excluded from analysis. PmWebSpec incorporates the recommended flags outlined by CDISC ADaM popPK IG, and users have the option to add their own flags if required (Fig. 3B). Additionally, this application enables a search function to locate flags used in similar projects previously.
Confirmations
The Confirmations section is designed to document additional information that is not captured in the dataset specification. This may include any email communications regarding the development of an algorithm or the confirmation of source data to select a certain variable for analysis. It helps trace back the logic of programming and can be beneficial for future projects.
Features of PmWebSpec
PmWebSpec serves two main functions: managing dataset specifications and offering tools to streamline the entire project lifecycle, from initial setup to completion. These functions are organized into eight features, which are accessible from the home page, facilitating navigation through the application (summarized in Table II).
Examples
To help users navigate PmWebSpec, we have provided several examples that cover the different features of the application. These examples include the development of dataset specifications, from creation to approval, preparing for e-Submission (e-Sub), downloading dataset specifications, generating SAS code, and modifying templates.
Example 1: Dataset Specification Lifecycle/Management
Step 1a: Create a Dataset Specification from the PPK-CDISC Template
To generate a new dataset specification using a pre-populated template, users can choose the “Create New” feature available on the home page. Users are prompted to select a template from the drop-down list.
Once the PPK-CDISC template is selected, the dataset specification page will appear, pre-populated with dataset attributes in the Specification Information, variables and their attributes in the Dataset Structure, and derivations and flags in the Derivations from the CDISC ADaM popPK IG.
Once users fill out the required information in the specification, they can submit it. Upon submission, it will be assigned a specification ID and labeled as version 1. The specification can be further revised, as needed, in “Modify” (step 2).
This feature is often used by pharmacometricians when working with a new compound, a new indication, or a new type of analysis, where no existing dataset specification is available.
Step 1b: Create a Dataset Specification from an Existing One
If there is already a similar specification available, the “Import Existing” feature can be used to create a new one. Users are directed to a page containing a set of filters and search results (all results are displayed, by default). Users can filter by specification ID, compound name, dataset type, created by, modified by, and indication to find the desired dataset specification (Fig. 4).
Once the specification ID is selected, users will be prompted to choose a version to proceed to the dataset specification. This page will have all the information pre-populated from the existing dataset specification, except for the project description and paths, as these details may not be the same. Users can make modifications as necessary to all sections of the specification, including modifications to the Dataset Structure table, shown in Fig. 2 and the Derivations table in Fig. 3. After completing and submitting, it will be assigned a specification ID and default to version 1.
The benefit of using this option is that it allows users to reuse a dataset specification that already exists for a similar analysis. This saves time and effort in customizing a new specification from scratch. This feature is particularly useful when pooling a new dataset with an existing one, as it ensures that both datasets have a similar dataset structure and are developed using the same rules.
Step 2: Modifying a Dataset Specification
To update a dataset specification, users can use the “Modify” feature. This feature will direct them to the same page as shown in Fig. 4, with the exception that the approved dataset specifications will not be displayed in the results. Users can use the same filters to select a specification and its version, which will lead them to the dataset specification.
When modifying a dataset specification, the page will appear similar to the one in step 1. However, there are a couple of differences. Firstly, the specification information section will include fields to record the changes made and the person who is making the change. Secondly, users have the option to save their progress, even if the page is only partially completed. It is important to note that when a dataset specification is being modified, it is locked to prevent other users from making changes simultaneously. This helps prevent any potential loss of information due to conflicts. The lock will be released when the dataset specification is submitted.
Users use this feature to update dataset specifications, including variables and their attributes and derivations. It is common that there are multiple updates to a dataset specification before finalizing it. This application maintains a version history of all modifications made to dataset specifications, ensuring transparency and traceability during the dataset specification development. It provides an option to retrieve previous versions if necessary, offering flexibility in managing the dataset specifications.
Step 3: Review/Approve a Dataset Specification
The “Review/Approve” feature provides functions that allow users to view the dataset specifications as a complete document, both during and after the dataset specification development. It is useful when users need to look up information or perform QC checks. Users can search for the dataset specification using the same filters mentioned in the previous steps. It opens an HTML page displaying all the contents from the dataset specification. Users also have the option to view it as a PDF document. Once the dataset specification is finalized, pharmacometricians can sign off on the document using the signature panel located at the bottom of the page. When the dataset specification is approved, no further modifications are allowed.
Reviewing and approving dataset specifications is crucial because it allows pharmacometricians and programmers to align on the final version of the specifications, considering various aspects of dataset creation such as source data usage, derivation methods, and imputation rules, prior to finalizing the dataset.
Example 2: Exporting a Dataset Specification for e-Sub Preparation
The “Export eSub” feature enables users to convert dataset specifications into eCTD compliant data definition file format including variable name, label, type, codes, and comments (7). To access this function, users can select the “Export eSub” feature and will be prompted to select a specification ID. The e-Sub dataset specification will be displayed on the page (Fig. 5). Within this page, users can update the dataset label, variable name, and attributes. Additionally, they can modify variable order or add/delete variables to match the dataset before exporting the data definition file.
Example 3: Downloading a Dataset Specification
Dataset specifications can be downloaded using the “Toolkit” feature on the home page. This will direct them to the same filters that were described earlier. Users can then choose the specification ID they desire and proceed to download the dataset specifications. Dataset specifications can be downloaded either locally to the desktop or to a server, in three formats: PDF, Word, and CSV. Dataset specifications in Word format can be appended to PMx reports, which help regulatory agencies in understanding the dataset creation process. PDF or Word dataset specifications can be shared with external partners for collaborations on dataset creation or analysis. Internally, we use the CSV dataset specifications to automate the QC process of the analysis dataset.
Example 4: Generating SAS Code from a Dataset Specification
The “Toolkit” feature includes an additional tool for automatically generating SAS code. Users can access this tool in the same manner as described in example 3. An example of SAS code is shown in Fig. 6.
During dataset preparation, programmers often spend significant time on tasks such as variable ordering and adding variables labels. This tool simplifies the process by extracting information from dataset specifications and generating SAS code. This code can be used to order variables, add variable labels, derive standard variables, round values, and impute missing values as necessary. By automating these tasks, programmers can save valuable time and focus on handling more complex algorithms and data issues. While the application currently provides SAS code, it can easily be translated to other programming languages. Additionally, future releases are planned to include the addition of R code.
Example 5: Modifying Built-in Templates and Derivations
The “Manage” feature includes a tool for template management. This application provides built-in templates that are designed to align with current practices. However, updates to the standards may be required to address study or project-specific issues. Maintaining up-to-date and user-friendly templates is crucial for all users. System administrators have the flexibility to modify these templates promptly after new standards become available, ensuring that new dataset specifications adhere to the latest standards without any delay.
To modify templates, system administrators can use the “Manage” feature and select “Modify Template”. Modifications can be made to existing flags and variables, such as adding or removing variables or flags, modifying the variable attributes and notes, and modifying notes and comments for flags. Users can also choose “Update Derivation” to add, remove, or modify derivation formulas.
Conclusion
Efforts have been made to standardize PMx datasets across the industry. In 2020, the International Society of Pharmacometrics (ISoP) Data Standards working group published dataset standards for popPK analysis (8) which set the ground for the CDISC ADaM popPK IG (4). PmWebSpec effectively implements the most recent standards in an automated way and ensures consistency in dataset specifications across projects, improving the quality of the dataset specifications and the analysis dataset.
PmWebSpec facilitates seamless sharing of the data across organizations and streamlines collaboration with external partners. The built-in templates eliminate the burden on pharmacometricians and programmers to manually populate all the standard variables, attributes, derivations, flags, and imputation rules. It also enables automation of data definition file for e-Sub and generation of SAS code to facilitate popPK dataset creation.
PmWebSpec serves as a central repository for all dataset specifications, for tracking, reusing, and referencing. To date, there are over 150 users and more than 580 dataset specifications that have been created in this application. This tool supports best practices in PMx and open innovation and its internal success indicates its potential for broader use across the PMx community. It is updated when there are changes to the standards or new features are incorporated. This tool can be expanded in the future to include additional functionalities in the dataset preparation workflow.
Additional Information
Design and Infrastructure of the Web Application
This user interface of this application is developed using Hypertext Preprocessor (PHP) v8.0 and deployed on the Amazon Web Services (AWS) platform. The application runs on AWS Elastic Beanstalk environment, and AWS Relational Database Service (RDS) with mySQL is used for storing application metadata and transactional data. There are two databases associated with this application: the template database, which is used to store dataset specification templates and user information, and the working database, which is used to store working specifications, metadata, and transactional data. Files, such as dataset specifications and attachments, generated by this application can be transferred to a local Linux server via AWS Simple Storage Service (S3) bucket.
Availability
This application is now available on GitHub (https://github.com/BMS-CPP/PMWebSpec) and is open to the public. A user manual is provided to help users in setting it up. This repository will be maintained by BMS CPP (Bristol Myers Squibb, Clinical Pharmacology and Pharmacometrics) and will be updated whenever a new release with enhancements is published.
References
Implementing Traceability in CDISC-Compliant Studies [Internet]. 2024. Available from: https://www.cdisc.org/video/traceability. Accessed 21 Feb 2024.
National Academies of Sciences, Engineering, and Medicine [Internet]. [cited 2024 Feb 21]. Reproducibility and replicability in science. Washington, DC: The National Academies Press. 2019 Available from: https://doi.org/10.17226/25303.
Thanneer N, Roy A, Sukumar P, Bandaru J, Carleen E. Best practices for preparation of pharmacometric analysis data sets. Poster session presented at: 5th American Conference on Pharmacometrics; 2014 Oct 12–15; Las Vegas, USA.
Basic Data Structure for ADaM popPK Implementation Guide v1.0 [Internet]. 2023. Available from: https://www.cdisc.org/standards/foundational/adam/basic-data-structure-adam-poppk-implementation-guide-v1-0. Accessed 21 Feb 2024.
Thanneer N, Chen L, Boyle B, Tang C, Dombrowsky E. Web app for creating pharmacometric analysis dataset specification form. Poster session presented at: 9th American Conference on Pharmacometrics; 2018 Oct 7–10; San Diego, USA.
Study Data Technical Conformance Guide [Internet]. 2023. Available from: https://www.fda.gov/media/153632/download. Accessed 21 Feb 2024.
Dombrowsky E, Sukumar P, Bandaru J, Roy A, Thanneer N. Best practices for preparation of submission quality data sets for pharmacometric analysis. Poster session presented at: 7th American Conference on Pharmacometrics; 2016 Oct 23–26; Bellevue, USA.
Basic Data Structure for Population Pharmacokinetic (popPK) Analysis [Internet]. 2020. Available from: https://go-isop.org/wp-content/uploads/2020/11/PopPK-Data-Standard-Implementation-Guide-1.pdf. Accessed 21 Feb 2024.
Acknowledgements
The authors would like to acknowledge the assistance provided by BMS research IT Team supporting Clinical Pharmacology and Pharmacometrics.
Funding
This work was sponsored and funded by Bristol Myers Squibb.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to conception and design of the work. L.C., E.D., and N.T. drafted the article. All authors took part in revising it critically for important intellectual content, agreed to submit to the current journal, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Consent for Publication
All the authors have reviewed and concurred with the manuscript.
Conflict of Interest
L.C., E.D., B.B., C.T., and N.T. are employees and hold equity ownership in Bristol Myers Squibb.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, L., Dombrowsky, E., Boyle, B. et al. PmWebSpec: An Application to Create and Manage CDISC-Compliant Pharmacometric Analysis Dataset Specifications. AAPS J 26, 39 (2024). https://doi.org/10.1208/s12248-024-00910-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-024-00910-0