Abstract
Background
Anthrax, a zoonotic disease caused by the spore-forming bacterium Bacillus anthracis, remains a major global public health concern, especially in countries with limited resources. Sierra Leone, a West African country historically plagued by anthrax, has almost been out of report on this disease in recent decades. In this study, we described a large-scale anthrax outbreak affecting both animals and humans and attempted to characterize the pathogen using molecular techniques.
Methods
The causative agent of the animal outbreak in Port Loko District, Sierra Leone, between March and May 2022 was identified using the nanopore sequencing technique. A nationwide active surveillance was implemented from May 2022 to June 2023 to monitor the occurrence of anthrax-specific symptoms in humans. Suspected cases were subsequently verified using quantitative polymerase chain reaction. Full-genome sequencing was accomplished by combining long-read and short-read sequencing methods. Subsequent phylogenetic analysis was performed based on the full-chromosome single nucleotide polymorphisms.
Results
The outbreak in Port Loko District, Sierra Leone, led to the death of 233 animals between March 26th and May 16th, 2022. We ruled out the initial suspicion of Anaplasma species and successfully identified B. anthracis as the causative agent of the outbreak. As a result of the government's prompt response, out of the 49 suspected human cases identified during the one-year active surveillance, only 6 human cases tested positive, all within the first month after the official declaration of the outbreak. The phylogenetic analysis indicated that the BaSL2022 isolate responsible for the outbreak was positioned in the A.Br.153 clade within the TransEuroAsian group of B. anthracis.
Conclusions
We successfully identified a large-scale anthrax outbreak in Sierra Leone. The causative isolate of B. anthracis, BaSL2022, phylogenetically bridged other lineages in A.Br.153 clade and neighboring genetic groups, A.Br.144 and A.Br.148, eventually confirming the spillover of anthrax from West Africa. Given the wide dissemination of B. anthracis spores, it is highly advisable to effectively monitor the potential reoccurrence of anthrax outbreaks and to launch campaigns to improve public awareness regarding anthrax in Sierra Leone.
Graphical Abstract

Similar content being viewed by others
Background
Anthrax is a zoonotic disease with a significant historical background. Despite being a long-standing issue, it remains a prominent global public health concern, particularly in resource-limited regions [1]. The disease, affecting both humans and animals, is caused by Bacillus anthracis, a gram-positive, spore-forming bacterium. Infection with B. anthracis typically results in cutaneous, gastrointestinal, inhalational, or injectional anthrax. Cutaneous anthrax is the prevailing form in humans, accounting for over 95% of reported human cases [1]. This form of anthrax typically has a comparatively lower mortality rate, whereas gastrointestinal and inhalational forms of anthrax, although less prevalent, are characterized by a significantly higher lethality rate and are considered the primary cause of most anthrax-related fatalities in humans [2, 3]. Unlike humans, both wild herbivores and livestock predominantly encounter acute and lethal consequences as a result of gastrointestinal exposure to spores while grazing [1, 4]. Livestock anthrax frequently serves as a source of secondary human infections, mainly due to the handling and consumption of animal products that are inadvertently contaminated [5, 6]. According to available data, approximately 1.83 billion individuals were considered to be living in areas at risk of anthrax infection [3]. According to estimates, approximately 1.1 billion livestock are susceptible to the disease, mainly in rural rainfed systems, with a particular emphasis on sub-Saharan Africa [1].
Africa is one of the most threatened continents, facing the challenge of anthrax outbreaks. These outbreaks have been extensively documented in multiple countries, affecting wildlife, domestic animals, and humans alike [1, 5,6,7,8]. However, it is believed that the actual number of cases is considerably underestimated due to inadequate diagnostic capabilities [8, 9]. Based on modeling techniques, previous studies have identified several regions in eastern, central, and southern Africa, as well as an east‒west corridor spanning from Ethiopia to Sierra Leone in the Sahel region, as having favorable conditions for the spread of B. anthracis [8, 10]. However, thus far, only Botswana, Zambia, Uganda, Tanzania, Namibia, South Africa, Nigeria, Cameroon, Chad, Ghana, and Ethiopia have documented verified instances of anthrax through the utilization of molecular and/or bacteriological techniques [10,11,12]. In Sierra Leone, sporadic instances of suspected anthrax can be identified in the annual reports of the Ministry of Agriculture between 1947 and 1994. The latest recorded outbreak data date back to 1994, during which 8 individuals lost their lives as a result of consuming deceased goat meat sourced from Guinea (Personal interview). Recently, there have been four reported cases of suspected cutaneous anthrax in human individuals within the vicinity of Kamakwie, Sierra Leone [13]. Regrettably, none of these records have been supported by dependable molecular or validated bacteriological methods. The absence of molecular verification significantly impedes the utility of these outbreak records as a reliable genetic resource for disease control.
B. anthracis is characterized by a relatively monomorphic nature, exhibiting a highly conserved genome composition. This implies that the genetic relatedness between descendant strains can be readily ascertained through the utilization of genomic molecular techniques, such as multilocus variable-number-of-tandem-repeats analysis (MLVA) and canonical single nucleotide polymorphisms (SNPs) [14, 15]. According to the analyses conducted using MLVA and the 12 canonical SNPs, B. anthracis exhibited a phylogenetic division into three primary branches, namely, A, B, and C. These branches were further classified into several sublineages, each characterized by distinct geographical distributions [14, 16]. With the emergence of cost-effective whole-genome sequencing methods, the field of phylogenetic reconstruction has witnessed a significant increase in the utilization of genome-wide SNPs as well as core-genome-based multilocus sequence typing (cgMLST) [16,17,18,19]. These methods offer improved strain clustering resolution, rendering them a potent tool for tracing outbreaks and modeling anthrax transmission [20,21,22]. Among the three primary phylogenetic branches of B. anthracis, isolates belonging to the B and C branches are exceedingly uncommon and geographically limited, often found only in specific locations or archival sources. In contrast, the A branch of B. anthracis exhibits a global distribution and accounts for more than 90% of outbreaks [14]. Within the A branch, the majority of the sublineages demonstrate a distinct geographical distribution. However, the TransEuroasian (TEA) sublineage has achieved significant global dissemination, possibly due to a rapid clonal radiation event within a short timeframe. This is evident from the remarkably short phylogenetic branches [21, 22]. In the West African region, there exists a limited number of publicly accessible complete genomes of B. anthracis, which were all isolated in Senegal and Gambia [23]. All these isolates belong to the TEA sublineage and exclusively form a specific subclade that has been recently designated as A.Br.148, according to the nomenclature of the full-chromosome SNP phylogeny system [19, 22]. Of note, despite the constraints of public genomic resources, West Africa bears a significant burden of anthrax caused by B. anthracis lineages that are primarily prevalent in this specific region [6, 9, 24]. A notable illustration is the exclusive presence of the specific lineage A.Br.148 in West Africa. This B. anthracis lineage, coupled with a distinct lineage found in Cameroon, Chad, Mali, and Nigeria that lacks spore-surface-associated anthrose [12, 25, 26], as well as the nontypical B. anthracis strains isolated from wild apes in Côte d'Ivoire and Cameron [27, 28], provides a glimpse into the complex nature of anthrax epidemiology in West Africa.
In this study, we provide a detailed account of an anthrax outbreak in Sierra Leone that exhibited an unprecedented magnitude. This outbreak led to the unfortunate demise of more than 200 animals and caused infections in six individuals. The analysis of the full-genome sequence indicated that the strain responsible for the outbreak was distinct from all existing West African lineages but may be associated with the anthrax outbreaks in London, Scotland, and New York between 2006 and 2008.
Methods
Sample collection during the outbreak and the study setting of the post-outbreak active surveillance
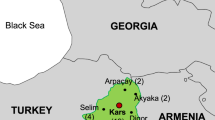
On March 26th, 2022, herbivorous livestock in Thinka Barreh village (8°48′32″N, 12°55′40″W) of the Bakeloko Chiefdom, located in the Port Loko District of Sierra Leone, were reported to be afflicted by an unidentified disease (Fig. 1). On April 11th, samples of tissue from the spleen and liver of a deceased bovine, along with blood samples from eight live bovines, were collected from the Affina Jalloh farm and submitted to the Tropical Infectious Diseases Prevention and Control Center of Sierra Leone (IDPC) for verification of the initial suspicion of Anaplasma infection, as reported by local veterinarian. On May 4th, a comprehensive collection of clinical samples was conducted, comprising 40 blood samples from live cattle and 6 blood samples from live sheep, as well as tissue samples from the heart, liver, spleen, lung, and kidney of a newly deceased sheep. These samples were collected from two neighboring farms, Chernaor Bah and Amadu Wurie Bah, where the grazing areas overlapped with the Affina Jalloh farm. After collection, the samples were promptly dispatched to the IDPC in a cold chain for the molecular identification of the causative agent.
The identification and official response to the anthrax outbreak in Port Loko District of Sierra Leone. The upper half of the diagram shows time points related to animal outbreaks in lilac, while the lower half depicts time points associated with human cases in cyan. IDPC The tropical Infectious Diseases Prevention & Control Center of Sierra Leone
As an effort to effectively control the outbreak, prompt implementation of active surveillance was carried out following its identification. Sierra Leone is a country with a population of approximately 7.54 million inhabitants. It is administratively divided into five Regions and sixteen Districts, which are further subdivided into Chiefdoms, followed by villages and communities. In the context of active surveillance, a comprehensive approach was employed to encompass all villages and communities across the entire nation. The active surveillance commenced on May 16th, 2022, and was concluded in June 2023, covering both the rainy and dry seasons.
Rapid identification of the causative agent responsible for the outbreak
All experimental procedures were conducted within a designated A2-type biological safety cabinet situated in a biosafety level 3 (BSL-3) facility. The liver sample underwent two rounds of rinsing with ethanol (70%) and was subsequently divided into smaller sections. The materials underwent a washing process using 1 ml of distilled water, and 200 μl of the supernatant was utilized for the extraction of DNA/RNA using the TGuid S32 magnetic DNA/RNA isolation kit (Tiangen, Beijing, China) following the manufacturer's instructions. The nucleic acid was sterilized at 95 °C for 20 min. The concentration of the nucleic acid was determined using a Qubit 4 fluorometer (Invitrogen, Carlsbad, USA). A nanopore sequencing library was prepared by utilizing the DNA samples and the Rapid Barcoding and Sequencing kits provided by Oxford Nanopore Technologies (Oxford, UK). The library was subsequently subjected to sequencing on a MinION Mk1B device using an R9.4.1 flow cell (Oxford Nanopore Technologies, Oxford, UK). During the one-hour sequencing process, the acquisition of raw data, base calling, and demultiplexing were carried out in real time using MinKNOW software associated with the device, utilizing a "superaccuracy" model. A swift annotation was then performed using the software Centrifuge (release 1.0.3) with the "Bacteria, Archaea, Viruses, Human (compressed)" database [29].
Confirmation of B. anthracis through quantitative PCR (qPCR) and cultivation
Tissue samples were processed according to the aforementioned procedure, with a minor adjustment. A homogenization process was performed on each tissue sample, where 200 mg of the sample was mixed with 1 ml of normal saline solution. A volume of 200 μl of the supernatant obtained from the tissue homogenate or blood sample was used for DNA extraction. The analysis of B. anthracis DNA was conducted through qPCR to examine the presence of chromosomal marker dhp61 (BA_5345) as well as plasmid markers pagA (pXO1) and capC (pXO2) using the primers as previously described [30, 31].
For cultivation, the liver tissue sample was subjected to two rounds of rinsing with ethanol (70%) and distilled water and subsequently partitioned into two distinct sections. The cutting plane was applied onto the nutrient agar, which was incubated at a temperature of 37 °C for 24 h. Colonies were subsequently purified using the streaking method. One colony was selected to prepare a slide for Gram staining.
Procedure of the active surveillance
Post-outbreak active surveillance of human anthrax was carried out in compliance with the directive of the Ministry of Health and Sanitation. Community health workers were given instructions to identify cases displaying typical symptoms indicative of suspected anthrax. These symptoms encompass the manifestation of a painless or itchy papule accompanied by excessive swelling, which subsequently progresses into a vesicular morphology, ruptures, and eventually forms an ulcer and black eschar [4]. If individuals exhibiting such symptoms had been in close proximity to, or had come into contact with, animal meat sourced from Port Loko District within a two-week period prior to the manifestation of symptoms, a swab specimen and a whole blood sample were collected. These samples were expeditiously transferred to the IDPC in a cold-chain system within 24 h. In the BSL-3 laboratory, the samples were subjected to analysis using qPCR and cultivation techniques, as previously stated. Samples that tested positive for both chromosome and plasmid markers were classified as confirmed cases.
Full-genome sequencing
Cells from the culture were harvested and digested with lysosome at 20 mg/ml and RNase A at 2 mg/ml for 1 h, followed by genomic DNA extraction using the TGuide Bacteria DNA Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. Sterilization of the resulting DNA was performed using 0.22-μm filters, which was further confirmed by culturing on nutrient agar. Eligible DNA with no positive colonies on the plate within 72 h was subsequently quantified using the Qubit 4 fluorometer and transferred out of the BSL-3 laboratory [17]. Full-genome sequencing was conducted by a combination of short-read sequencing and long-read sequencing techniques [32, 33]. For long-read sequencing, a nanopore sequencing library was prepared employing the Ligation Sequencing Kit and the Native Barcoding Kit (Oxford Nanopore Technologies, Oxford, UK). The library was sequenced on a MinION Mk1B device for 24 h using an R9.4.1 flow cell (Oxford Nanopore Technologies, Oxford, UK) following the manufacturer's manuals. Base calling, demultiplexing, and de novo assembly of the long-read sequencing data were performed using the corresponding pipelines and tools in EPI2ME application v5.1.1 (Oxford Nanopore Technologies, Oxford, UK) [17].
Short-read sequencing was performed on an Illumina MiSeq platform using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, USA) and the MiSeq Reagent Kit v3 (2 × 300 bp) chemistry (Illumina, San Diego, USA). High-quality paired-end reads were subjected to de novo assembly into contigs in SPAades 3.11.1 [34] prior to further refining by SNP and indel correction using SAMtools 1.7 and Pilon 1.22 [35, 36]. The assembly and polishing of the combined long reads and short reads data were ultimately conducted using MicroPIPE v0.9 [37]. The primary data generated in this study were deposited in the NCBI Sequence Read Archive (SRA) repository under the BioProject number PRJNA875505.
Full-chromosome SNP calling and phylogenetic construction
To determine the phylogenetic position of the B. anthracis strain isolated in this outbreak, we retrieved 236 representative full genome sequences that were used in the initial global phylogenetic reconstruction [22] and the most recent molecular typing practice based on full-chromosome SNPs of B. anthracis [32]. The Parsnp tool from the Harvest Suite [38] was then used for the core chromosome multiple alignment with the ‘Ames Ancestor’ (NC_007530.2) stain serving as the reference chromosome. Chromosome-wide core SNPs were called and exported as concatenated SNP sequences. HarvestTools v1.2 from the same suite was employed to generate a variant calling file (vcf) listing all SNP-positions [32, 39]. Adjacent SNP positions, as well as sites with unknown nucleotides (N), were manually removed in the vcf for the purpose of enhancing data quality [39]. According to this curated vcf, the concatenated SNP sequences were renewed using the HarvestTool v1.2. These new SNP sequences were employed to infer the phylogenetic trees with the maximum likelihood model in RAxML v8.2.12 [40]. Phylogenetic trees were annotated using the online iTol platform [41]. A minimum spanning tree was calculated in Grapetree [42] with the curated vcf (binary format) as an input [42].
Results
Identification and confirmation of the anthrax outbreak in Sierra Leone
The salient aspects of the outbreak are illustrated in Fig. 1. The diseased animals exhibited a range of symptoms including weakness, depression, shivering, mortality, and postmortem hemorrhage from the oral and nasal cavities (Fig. 2A). Local veterinarians hypothesized that the outbreak could potentially be attributed to a severe infection caused by Anaplasma species, given the observation of leukemia cases in cattle on the same farm. In the BSL-3 laboratory, a PCR-based screening was conducted on the bovine samples collected on April 11th, 2022 to identify the presence of the Anaplasma species [43]. The screening revealed that all samples yielded negative results, thereby refuting the initial hypothesis.
Anthrax broke out in animals in Port Loko District and spread across the country to humans. A The head of a diseased cow at site of death with postmortem hemorrhage from the oral and nasal cavities. B Gram staining of a culture taken from the liver of a sheep deceased during the outbreak (magnification 400 ×). C The geographical locations of the outbreak and cases. The red star marks the location of the initial animal outbreaks. The circles represent the locations of the Freetown case (left), the Karene cases (middle), and the Kailahun case (right). D, E Cases of human anthrax infections from Karene (red arrow)
On May 3rd, the Department of Animal Sciences of Njala University led a team to the outbreak area in attempt to identify the etiology of the ongoing outbreak, as the disease's rapid dissemination resulted in substantial livestock mortality, leading to significant economic losses for farmers. Following the collection of samples, we promptly conducted an identification of the potent causative agent utilizing a random long-read sequencing technique, which has been demonstrated to be efficacious in emergency preparedness [17]. The results revealed a significant presence of anthrax infection in the deceased sheep, as the majority of the sequencing data from this sample were identified as originating from B. anthracis or closely related Bacillus cereus and Bacillus thuringiensis, with the exception of host sequences (Appendix, Table S1). Given the inherent error-prone nature of long-read sequencing, we conducted additional validation of the results by employing the qPCR method to assess the presence of the chromosomal marker dhp61 (BA_5345), as well as the plasmid markers pagA (pXO1) and capC (pXO2) of B. anthracis in the samples. It was not surprising that the DNA samples collected from the spleen, liver, kidney, and lung all yielded strong positive results, as evidenced by cycle threshold (Ct) values ranging from 14 to 22. In contrast, all samples obtained from live animals tested negative for B. anthracis genes. These observations clearly indicated that when considering the Ct value of the positive control (which was 31), there was a very high bacterial load in the tissue sample of the sheep that died in the outbreak. In addition, a qPCR analysis was conducted on the bovine samples used for Anaplasma screening, yielding comparable outcomes. The analysis of the tissue samples exhibited a positive curve with low Ct values (ranging from 15–20), indicating the significant presence of B. anthracis. Conversely, the blood samples obtained from live animals tested negative for B. anthracis. Additional isolation and purification of B. anthracis from the liver and spleen tissues of the deceased sheep were performed through culturing techniques. Gram staining of the colony revealed a unique morphology characterized by elongated and rod-shaped structures, displaying a gram-positive appearance (Fig. 2B). The aforementioned findings provided compelling evidence supporting the assertion that the outbreak was indeed a result of a B. anthracis infection.
As of May 9th, the Affina Jalloh farm reported that 66.7% (14 out of 21) of the cattle, 100% (5 out of 5) of the sheep, and 100% (4 out of 4) of the goats had been affected by the disease. Similarly, the Chernor Bah farm reported that 31 out of 85 cattle (36.5%), 18 out of 20 sheep (90%), and 16 out of 16 goats (100%) had also perished due to the disease. These numbers signified a substantial outbreak. In contrast, in an effort to mitigate the economic losses, the farmers sold the meat of the animals that succumbed to the outbreak in the market. This action led to the extensive spread of the threat, which originated from the Port Loko District and had implications for the entire nation, affecting both animal and human populations. Consequently, the findings were promptly communicated to the Ministry of Agriculture.
The official response and active surveillance
On May 16th, 2022, the Ministry of Agriculture and the Ministry of Health and Sanitation collaboratively announced the official declaration of an anthrax outbreak to effectively mitigate and reduce the risk of transmission associated with the emergency. Subsequently, a comprehensive set of control measures was implemented, encompassing stringent regulations that govern the production, processing, and marketing of livestock and livestock products. As of the declaration of the outbreak, a cumulative number of 233 livestock (91 cattle, 53 goats, and 79 sheep) had been reported as deceased in the impacted region, with a significant portion of the meat having entered the market. The Ministry of Health and Sanitation expeditiously implemented an enhanced surveillance program for anthrax within the local communities. On May 16th, a total of four samples (consisting of three swabs and one blood sample) were collected from seven suspected human cases in a village located in the Karene District (Fig. 2C) and subsequently submitted to the IDPC, where all three swabs tested positive for B. anthracis by qPCR. All three confirmed cases exhibited a distinctive symptom of cutaneous anthrax (Fig. 2D, E) and had been in close proximity to deceased livestock within two weeks. Between May 19th and June 17th, an additional three cases of cutaneous anthrax were confirmed in the IDPC. These cases were reported in the Districts of Karene, Freetown, and Kailahun, as shown in Fig. 2C. All three cases had a history of contacting animal meat within a week prior to the onset of symptoms. Since June 2022, the Ministry of Health and Sanitation has implemented an ongoing active surveillance program to monitor the occurrence of anthrax and other diseases characterized by skin lesions. Over a twelve-month duration that included both a rainy season and a dry season, a total of 43 samples obtained from individuals with suspected cases were subjected to testing for anthrax using both qPCR and cultivation methods [4, 11]. The results of these tests revealed that none of the 43 samples exhibited any traces of anthrax.
The phylogenetic placement of the B. anthracis strain responsible for the outbreak
From the tissue samples obtained from deceased cattle and sheep, we successfully isolated and purified two strains of B. anthracis. The integration of long-read sequencing data and short-read sequencing data derived from the genomic DNA sample [37] resulted in the identification of three expected contigs, encompassing the complete genome as well as two virulent plasmids, pXO1 and pXO2. The average sequencing depth achieved in this study was 124-fold. The application of short-read sequencing data for polishing purposes effectively mitigated the indel-introducing effect commonly associated with the long-read sequencing technique [37], thereby exhibiting a high level of reliability [17, 32]. The full-chromosome SNP sequences were identical between the two isolates. Consequently, considering the shared origin of these samples from a single outbreak, we categorized them as a unified strain, denoted as BaSL2022 (Bacillus Sierra Leone 2022). The construction of the global dendrogram of the B. anthracis phylogeny involving the analysis of 236 representative genomes and BaSL2022 was conducted using the maximum likelihood method [14], which positioned BaSL2022 within the A.Br.008/009 (TEA) sublineage (Fig. 3A) of the A branch. The precise phylogenetic placement of BaSL2022 within the TEA group is illustrated in Fig. 3B, which was generated using 72 representative genomes from the TEA lineage. BaSL2022 was classified within the canonical SNP group A.Br.011/009, more specifically falling under the A.Br.153 subgroup, which was strongly supported by bootstrapping analysis.
BaSL2022 was placed into A.Br.153 clade within the TranEuroAsian genetic group based on core-genome SNP analysis. A Global distribution of BaSL2022 shown in a maximum likelihood tree. The phylogenetic tree was constructed using core-chromosome SNPs of the representative genomes described previously [22, 32, 39]. Lineages are categorized and labeled into traditional groups. The TEA group, including BaSL2022, is highlighted in red. B The specific position of BaSL2022 within the TEA group. The phylogenetic tree was constructed using the maximum likelihood model. It included 72 representative genomes from the TEA group, as well as BaSL2022, with the reference strain being Ames and serving as the outgroup. The tree was drawn to scale, accurately representing the length of the branches. Nodes with a bootstrapping value above 90 are indicated by a black circle, while those between 70 and 90 are indicated by a gray circle. The lineages are displayed with the isolate's name, country of isolation, and year of isolation. The color blocks on the leaves represent the genetic subgroups of the TEA group, and the nicknames of these subgroups are also displayed in colored blocks outside the lineages. The outer layer displays the canonical SNP groups of isolates within the TEA group using circular blocks. The BaSL2022 is marked with star and shown in blue
Based on the core-chromosome SNPs of the isolates belonging to A.Br.153, A.Br.148, and A.Br.144, a minimum spanning tree was calculated and showed that BaSL2022 played a crucial role in connecting the evolutionary path from A.Br.144 (Fig. 4). The closest SNP distance observed between the A.Br.153 subclade and previously identified subclades was 175, specifically, between isolate BaSL2022 and Pollino (A.Br.144). The A.Br.153 subclade is exclusively characterized by lineages that are linked to the outbreaks in London, Scotland, New York, and Port Loko (Sierra Leone). Among the lineages implicated in these four outbreaks, the lineages associated with the London outbreak formed a distinct cluster and exhibited a closest genetic distance of 77 SNPs to BaSL2022. The Scotland isolate exhibited a direct genetic connection to BaSL2022, with a difference of 41 SNPs (Fig. 4).
BaSL2022 phylogenetically bridged A.Br.153 subclade and the neighboring lineages. Minimum spanning tree was calculated using the core-chromosome SNP profile by Grapetree [42]. Numerical SNP distances between the chromosomes of isolates in A.Br.153, A.Br.148, and A.Br.144 are depicted on the branches. The BaSL2022 is displayed in bright red. Outbreaks associated with the isolates in A.Br.153 are also presented
Discussion
Despite the extensive documentation of anthrax in West Africa, there exists a notable dearth of comprehensive data, particularly pertaining to the diagnosis and genetic sequences of B. anthracis, within this region. In the present investigation, we successfully identified an anthrax outbreak impacting both animals and humans in Sierra Leone through the utilization of sequencing and molecular techniques. This advancement signifies a noteworthy achievement in the realm of anthrax prevention and control in Sierra Leone.
In this anthrax outbreak, the mortality of 233 animals in Sierra Leone represents the highest number ever reported in Sierra Leone. Although laboratory confirmation was not conducted for the majority of the animal cases, the confirmation of anthrax infection in the sequentially deceased bovine and sheep, which were determined to be caused by the same strain, provides strong evidence to suggest that the suspected animal cases succumbed to anthrax exposure. From 1947 to 1994, Sierra Leone witnessed a succession of outbreaks that impacted both livestock and humans, as documented in the annual reports of the Ministry of Agriculture, with a particular focus on the Kamakwie area situated in the Northern Region. However, all these instances were diagnosed based solely on symptoms and, at most, with the assistance of microscopy [44]. This situation arose as a result of inadequate laboratory capacities and facilities, which were further compromised during the onset of the civil war in 1991. Such a predicament could potentially provide an explanation for the absence of any officially recorded instances of anthrax cases in the country since 1994. The scarcity of publicly accessible data on the disease further posed a potential contributing factor to the underdiagnosis and misdiagnosis of the disease [7, 44, 45]. The misdiagnosis also occurred at the beginning of the investigation of this anthrax outbreak, which notably hampers the timeliness of the implementation of intervention measures. In view of this, the identification of a widespread outbreak by employing nanopore sequencing to examine clinical samples of an unidentified ailment [17], as demonstrated in this study where we rapidly determined the causative agent without any preconceived notions, provides significant insights for tackling outbreaks of anthrax and other neglected diseases in Sierra Leone and similar countries.
It is worth mentioning that in 2018, a documented report was published regarding four suspected cases of cutaneous anthrax originating from the Kamakwie region of Sierra Leone [13]. In the report, the diagnosis of all cases was conducted using clinical interviews and enzyme-linked immunosorbent assays [13]. However, the application of this assay for diagnosing anthrax has not been widely endorsed due to its limited accuracy in this particular context [44]. This aspect holds particular significance when considering that the cases under investigation occurred in Kamakwie, an area where anthrax was endemic. Consequently, the exclusion of historical anthrax infection in these cases poses a significant challenge, as it may result in misleading outcomes in antibody-based diagnostic procedures. While the diagnosis of anthrax and similar diseases in Sierra Leone and neighboring countries has been impacted by various challenges [11], this paper presents the first report analyzing B. anthracis isolates and clinical specimens from suspected cases of cutaneous anthrax using the qPCR method, which has recently become more accessible and feasible in resource-limited countries due to capacity-building initiatives [11].
The prompt and proactive response from the Ministry of Agriculture and the Ministry of Health and Sanitation to the outbreak played a crucial role in effectively mitigating the risk of spreading. As evidenced by previous investigations [46] and the findings in this study, only six cutaneous anthrax cases were confirmed, all within a month after the implementation of intervention measures. The confirmation of human cases was achieved through qPCR, although attempts to culture B. anthracis were unsuccessful. This failure can be attributed to the presence of other bacterial species in the samples, as B. anthracis can be readily outcompeted [44]. Consequently, the utilization of molecular techniques for the purpose of tracking human cases presented notable difficulties, thereby raising the question of whether the occurrence of these cases was a consequence of secondary infection stemming from the animal outbreak [6] or if they constituted separate outbreaks. The results obtained from the subsequent long-term active surveillance indicated that the confirmed human cases were predominantly clustered within a limited time period after the outbreak, as opposed to being spread out over the entire duration of the surveillance. This finding provides support for the hypothesis that the human cases resulted from the animal outbreak rather than occurring independently. Notably, despite the absence of confirmed cases during post-outbreak surveillance, it is imperative to avoid underestimating the risk of future anthrax outbreaks, given the prolonged viability of B. anthracis spores in the environment, which can persist for several decades [2, 9, 22]. When taking into account the circumstances in which the processing and consumption of contaminated meat occurred locally before the outbreak was officially declared, it is crucial to express considerable concern regarding the repeated occurrence of this disease in Sierra Leone.
The BaSL2022 strain represents the initial identification of the B. anthracis isolate discovered in Sierra Leone. The analysis of full-chromosome SNPs revealed that BaSL2022 was classified within the A.Br.153 phylogenetic group and exhibited a close relationship with isolates (Fig. 3B, Fig. 4) obtained from three outbreaks related to drum-making and drumming activities [47] in Scotland (Scotland_484, 2006) [20, 47], London (London_493, 2008) [48], and New York (A3802, 2006) [20, 49]. The animal skins and hides used for the drums associated with the Scottland and New York cases were believed to have been imported from Guinea [20] and Cote d'Ivoire [49], respectively. The London cases were investigated in connection with animal hide sourced from various origins, including Gambia [48]. The observation that the genomes of the strains isolated from the outbreaks were largely similar to each other and constituted a distinct phylogenetic clade (A.Br.153) separate from other existing clades in the TEA lineage (Fig. 3B) further supported the inference that these strains, obtained from cases associated with animal-hide drums, likely originated from the same geographical region, most likely West Africa [20]. However, the verification of this hypothesis was pending, as the three sequences, which were obtained at a later time from samples collected in 2010 in West African Senegal and, specifically, Gambia [23], were found to be different from the sequences obtained from the cases in Scotland, London, and New York. In contrast, a distinct clade known as A.Br.148 [20] was formed, as depicted in Fig. 3B. In light of the aforementioned evidence, our findings present strong support for the conclusive element of the hypothesis, suggesting that the anthrax incidents in the United Kingdom and United States during the 2000s were caused by the spillover of B. anthracis from West Africa.
Considering the geographical origins of the hides associated with the outbreaks in Scotland (Guinea), New York (Cote d'Ivoire), and London (multiple origins including Gambia), the identification of BaSL2022 in Sierra Leone has revealed the potential for long-term endemicity of A.Br.153 in at least the west coast of West Africa. Furthermore, it is important to note that all the strains identified in West Africa were found to be genetically distinct from other phylogenetic groups worldwide [12, 23]. Therefore, the association of BaSL2022 with other A.Br.153 isolates signifies the first instance of B. anthracis lineages being transmitted from this specific geographical area to other continents.
Our study was subject to certain limitations. Firstly, high-throughput sequencing was not conducted during the initial detection of the outbreak. If a comprehensive analysis had been conducted, it is possible that the outbreak could have been detected and addressed more promptly, resulting in a reduced impact on farmers and lower costs associated with outbreak control. Secondly, the utilization of nanopore sequencing played a pivotal role in the identification of this outbreak. However, the efficiency of nanopore sequencing in RNA sequencing, as opposed to DNA sequencing, is a significant factor to consider. This is particularly true when taking into account the drastic manner in which we processed the samples, which ultimately limited the potential application of our procedure in other situations. Lastly, our attempts to isolate a strain of B. anthracis from the samples obtained from human cases were unsuccessful. Therefore, our conclusion regarding the human cases being a result of the animal outbreak was solely derived from the temporal distribution of human cases, without the presence of molecular evidence.
Conclusions
In the current investigation, the causative agent responsible for a widespread outbreak was successfully identified as B. anthracis through the utilization of the nanopore sequencing technique. The initial application of qPCR for the diagnosis of anthrax in this nation resulted in the confirmation of six human cutaneous anthrax cases among 49 suspected cases following the animal anthrax outbreak. We successfully isolated and purified the first Sierra Leonean B. anthracis strain BaSL2022 from the outbreak. Phylogenetic analysis revealed that this strain is distinct from the currently available West African lineages and belongs to the A.Br.153 clade. This clade exclusively includes isolates obtained from animal-hide-associated cases in the United Kingdom and United States between 2006 and 2008. This finding provides strong evidence to support the conclusive aspect of the hypothesis that the cases in the United Kingdom and United States were a result of the spillover of B. anthracis from West Africa.
In the present circumstances, the lack of awareness about anthrax greatly affected the initial diagnosis and delayed the timely control of the outbreak. Therefore, we propose that governmental bodies and scientific societies take the initiative to launch an intensified educational campaign aimed at raising awareness about anthrax, along with other neglected zoonotic diseases, among farmers, veterinarians, and disease surveillance personnel in Sierra Leone and other similar nations. Furthermore, given the extensive dissemination of contaminated animal meat throughout this outbreak, it is highly advisable to establish a comprehensive and ongoing surveillance system to effectively monitor the potential reoccurrence of anthrax outbreaks as the longevity of B. anthracis spores in the environment can span several decades.
Availability of data and materials
The sequencing data were submitted to the NCBI Sequence Read Archive under the BioProject number PRJNA875505. All the other data yielded in this study are shown in the paper as well as the supplementary materials.
Abbreviations
- cgMLST:
-
Core genome multilocus sequence typing
- Ct:
-
Cycle threshold
- MLVA:
-
Multilocus variable-number-of-tandem-repeat analysis
- qPCR:
-
Quantitative polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
- TEA:
-
TransEuroAsian
References
Carlson CJ, Kracalik IT, Ross N, Alexander KA, Hugh-Jones ME, Fegan M, et al. The global distribution of Bacillus anthracis and associated anthrax risk to humans, livestock and wildlife. Nat Microbiol. 2019;4(8):1337–43.
Carlson CJ, Getz WM, Kausrud KL, Cizauskas CA, Blackburn JK, Bustos Carrillo FA, et al. Spores and soil from six sides: interdisciplinarity and the environmental biology of anthrax (Bacillus anthracis). Biol Rev Camb Philos Soc. 2018;93(4):1813–31.
Swartz MN. Recognition and management of anthrax–an update. N Engl J Med. 2001;345(22):1621–6.
Sweeney DA, Hicks CW, Cui X, Li Y, Eichacker PQ. Anthrax infection. Am J Respir Crit Care Med. 2011;184(12):1333–41.
Aceng FL, Ario AR, Alitubeera PH, Neckyon MM, Kadobera D, Sekamatte M, et al. Cutaneous anthrax associated with handling carcasses of animals that died suddenly of unknown cause: Arua District, Uganda, January 2015-August 2017. PLoS Negl Trop Dis. 2021;15(8): e0009645.
Blackburn JK, Kenu E, Asiedu-Bekoe F, Sarkodie B, Kracalik IT, Bower WA, et al. High case-fatality rate for human anthrax, Northern Ghana, 2005–2016. Emerg Infect Dis. 2021;27(4):1216–9.
Dhliwayo TH, Chonzi P, Madembo C, Juru TP, Chadambuka A, Gombe NT, et al. Anthrax outbreak investigation in Tengwe, Mashonaland West Province, Zimbabwe, 2022. PLoS ONE. 2022;17(12): e0278537.
Otieno FT, Gachohi J, Gikuma-Njuru P, Kariuki P, Oyas H, Canfield SA, et al. Modeling the spatial distribution of anthrax in southern Kenya. PLoS Negl Trop Dis. 2021;15(3): e0009301.
Nana SD, Caffin JH, Duboz R, Antoine-Moussiaux N, Binot A, Diagbouga PS, et al. Towards an integrated surveillance of zoonotic diseases in Burkina Faso: the case of anthrax. BMC Public Health. 2022;22(1):1535.
Romero-Alvarez D, Peterson AT, Salzer JS, Pittiglio C, Shadomy S, Traxler R, et al. Potential distributions of Bacillus anthracis and Bacillus cereus biovar anthracis causing anthrax in Africa. PLoS Negl Trop Dis. 2020;14(3): e0008131.
Ashenefe Wassie B, Fantaw S, Mekonene Y, Teshale AM, Yitagesu Y, Tsige E, et al. First PCR confirmed anthrax outbreaks in Ethiopia-Amhara region, 2018–2019. PLoS Negl Trop Dis. 2022;16(2): e0010181.
Blackburn JK, Odugbo MO, Van Ert M, O’Shea B, Mullins J, Perrenten V, et al. Bacillus anthracis diversity and geographic potential across Nigeria, Cameroon and Chad: further support of a novel West African lineage. PLoS Negl Trop Dis. 2015;9(8): e0003931.
Adeleke AA, Fasomoyin OO, Adekunle KA. Cutaneous anthrax in Kamakwie, Karene District, Northern Sierra Leone. J Case Report: Clin Med. 2020;3(1):145.
Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J, et al. Global genetic population structure of Bacillus anthracis. PLoS ONE. 2007;2(5): e461.
Lista F, Faggioni G, Valjevac S, Ciammaruconi A, Vaissaire J, le Doujet C, et al. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 2006;6:33.
Bruce SA, Schiraldi NJ, Kamath PL, Easterday WR, Turner WC. A classification framework for Bacillus anthracis defined by global genomic structure. Evol Appl. 2020;13(5):935–44.
McLaughlin HP, Bugrysheva JV, Conley AB, Gulvik CA, Cherney B, Kolton CB, et al. Rapid Nanopore whole genome sequencing for anthrax emergency preparedness. Emerg Infect Dis. 2020;26(2):358–61.
Pearson T, Busch JD, Ravel J, Read TD, Rhoton SD, U’Ren JM, et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci USA. 2004;101(37):13536–41.
Abdel-Glil MY, Chiaverini A, Garofolo G, Fasanella A, Parisi A, Harmsen D, et al. A whole-genome-based gene-by-gene typing system for standardized high-resolution strain typing of Bacillus anthracis. J Clin Microbiol. 2021;59(7): e0288920.
Pullan ST, Pearson TR, Latham J, Mason J, Atkinson B, Silman NJ, et al. Whole-genome sequencing investigation of animal-skin-drum-associated UK anthrax cases reveals evidence of mixed populations and relatedness to a US case. Microb Genom. 2015;1(5): e000039.
Bassy O, Antwerpen M, Ortega-Garcia MV, Ortega-Sanchez MJ, Bouzada JA, Cabria-Ramos JC, et al. Spanish outbreak isolates bridge phylogenies of European and American Bacillus anthracis. Microorganisms. 2023;11(4):889.
Sahl JW, Pearson T, Okinaka R, Schupp JM, Gillece JD, Heaton H, et al. A Bacillus anthracis genome sequence from the Sverdlovsk 1979 autopsy specimens. MBio. 2016;7(5):e01501-e1516.
Rouli L, Robert C, Ndiaye M, La Scola B, Raoult D. Genomic analysis of three African strains of Bacillus anthracis demonstrates that they are part of the clonal expansion of an exclusively pathogenic bacterium. New Microbes New Infect. 2014;2(6):161–9.
Pilo P, Rossano A, Bamamga H, Abdoulkadiri S, Perreten V, Frey J. Bovine Bacillus anthracis in Cameroon. Appl Environ Microb. 2011;77(16):5818–21.
Maho A, Rossano A, Hachler H, Holzer A, Schelling E, Zinsstag J, et al. Antibiotic susceptibility and molecular diversity of Bacillus anthracis strains in Chad: detection of a new phylogenetic subgroup. J Clin Microbiol. 2006;44(9):3422–5.
Tamborrini M, Bauer M, Bolz M, Maho A, Oberli MA, Werz DB, et al. Identification of an African Bacillus anthracis lineage that lacks expression of the spore surface-associated anthrose-containing oligosaccharide. J Bacteriol. 2011;193(14):3506–11.
Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, Jacob D, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d’Ivoire and Cameroon. J Bacteriol. 2006;188(15):5333–44.
Leendertz FH, Yumlu S, Pauli G, Boesch C, Couacy-Hymann E, Vigilant L, et al. A new Bacillus anthracis found in wild chimpanzees and a gorilla from west and central Africa. PLoS Pathog. 2006;2(1):1–4.
Centrifuge [Available from: http://ccb.jhu.edu/software/centrifuge/. Accessed 15 Jul 2023.
Ellerbrok H, Nattermann H, Ozel M, Beutin L, Appel B, Pauli G. Rapid and sensitive identification of pathogenic and apathogenic Bacillus anthracis by real-time PCR. FEMS Microbiol Lett. 2002;214(1):51–9.
Antwerpen MH, Zimmermann P, Bewley K, Frangoulidis D, Meyer H. Real-time PCR system targeting a chromosomal marker specific for Bacillus anthracis. Mol Cell Probe. 2008;22(5–6):313–5.
Brangsch H, Golovko A, Pinchuk N, Deriabin O, Kyselova T, Linde J, et al. Molecular typing of Ukrainian Bacillus anthracis strains by combining whole-genome sequencing techniques. Microorganisms. 2022;10(2):461.
Lienemann T, Beyer W, Pelkola K, Rossow H, Rehn A, Antwerpen M, et al. Genotyping and phylogenetic placement of Bacillus anthracis isolates from Finland, a country with rare anthrax cases. BMC Microbiol. 2018;18(1):102.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11): e112963.
Murigneux V, Roberts LW, Forde BM, Phan MD, Nhu NTK, Irwin AD, et al. MicroPIPE: validating an end-to-end workflow for high-quality complete bacterial genome construction. BMC Genomics. 2021;22(1):474.
Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524.
Antwerpen M, Beyer W, Bassy O, Ortega-Garcia MV, Cabria-Ramos JC, Grass G, et al. Phylogenetic placement of isolates within the trans-Eurasian Clade A.Br.008/009 of Bacillus anthracis. Microorganisms. 2019;7(12):689.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6.
Zhou ZM, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28(9):1395–404.
Wei F, Song M, Liu H, Wang B, Wang S, Wang Z, et al. Molecular detection and characterization of zoonotic and veterinary pathogens in ticks from Northeastern China. Front Microbiol. 2016;7:1913.
Aminu OR, Lembo T, Zadoks RN, Biek R, Lewis S, Kiwelu I, et al. Practical and effective diagnosis of animal anthrax in endemic low-resource settings. PLoS Negl Trop Dis. 2020;14(9): e0008655.
Molyneux D, Hallaj Z, Keusch GT, McManus DP, Ngowi H, Cleaveland S, et al. Zoonoses and marginalised infectious diseases of poverty: Where do we stand? Parasit Vectors. 2011;4:106.
Wondmnew T, Asrade B. Case-control study of human anthrax outbreak investigation in farta woreda, South Gondar, Northwest Ethiopia. BMC Infect Dis. 2023;23(1):167.
Bennett E, Hall IM, Pottage T, Silman NJ, Bennett AM. Drumming-associated anthrax incidents: exposures to low levels of indoor environmental contamination. Epidemiol Infect. 2018;146(12):1519–25.
Anaraki S, Addiman S, Nixon G, Krahe D, Ghosh R, Brooks T, et al. Investigations and control measures following a case of inhalation anthrax in East London in a drum maker and drummer, October 2008. Euro Surveill. 2008;13(51):19076.
CDC. Inhalation anthrax associated with dried animal hides—Pennsylvania and New York City, 2006. MMWR Morb Mortal Wkly Rep. 2006;55(10):280–2.
Acknowledgements
We would like to extend our utmost gratitude to the personnel of the Department of Animal Sciences at Njala University for their unwavering commitment in the field, as they diligently worked to identify and verify this significant outbreak. The dedication of individuals involved played a crucial role in mitigating the impact of the outbreak. We would like to express our gratitude to Dr. Yajun Song from the Beijing Institute of Microbiology and Epidemiology, Dr. Lingwei Zhu from the Changchun Veterinary Research Institute, and Dr. Lifeng Zhao from Jilin Agricultural Science and Technology University for their invaluable contributions in providing diagnosis consultation and technical support. We would like to extend our gratitude to the 2022 Group of China CDC in Sierra Leone led by Dr. Canjun Zheng (China CDC) for their invaluable technical support in the field of short-read sequencing.
Funding
This work was supported by the tropical Infectious Diseases Prevention & Control Center of Sierra Leone.
Author information
Authors and Affiliations
Contributions
SW and ZM performed the identification, isolation, and sequencing of the pathogen. RS, AFS, MNK, and MES collected and processed the samples. DH, JSS, and MAV performed the active surveillance and human sample management. SW, BJ, YX, YZ, MZ, and ZM performed the laboratory detection of active surveillance. SW analyzed the data and drafted the manuscript. SW, MBJ, FS, SZ, RH, and ZM revised and edited the manuscript. All authors have read and approved the submitted version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This investigation was performed in response to an emergency and thus not considered purely as scientific research. The animal samples were collected upon request for diagnosis, which was approved by Njala University. The sampling of suspected human cases was approved by the Ministry of Health and Sanitation under an emergency mechanism. All patients or their guardians provided verbal consent to the collection of their samples. Case information other than the location and results was not included in the text.
Competing interests
The authors declare that there are no conflicts of interest.
Supplementary Information
Additional file 1: Table S1:
Annotation of reads obtained from the rapid identification.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, S., Suluku, R., Jalloh, M.B. et al. Molecular characterization of an outbreak-involved Bacillus anthracis strain confirms the spillover of anthrax from West Africa. Infect Dis Poverty 13, 6 (2024). https://doi.org/10.1186/s40249-023-01172-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40249-023-01172-2