Abstract
Background
Cystic echinococcosis (CE), caused by the larval stage of the complex Echinococcus granulosus sensu lato (s.l.), is a zoonotic parasitic disease with a high social burden in China. E. ortleppi is a species (formerly genotype 5 of E. granulosus s.l.) with unique epidemic areas (tropical areas), transmission patterns (mainly cattle origin), and pathological characteristics (large and small hook lengths) compared to other species that cause CE. A 19-year-old female patient in an area with no history of echinococcosis in Guizhou Province, China, was diagnosed with E. ortleppi infection in 2019. This study is to understand the source of this human E. ortleppi infection.
Methods
We performed computer tomography (CT) scans, surgical operation, morphological sectioning, molecular diagnosis, phylogenetic analyses, and epidemiological investigation in Anshun City, Guizhou Province, China in 2019.
Results
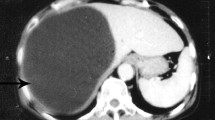
The patient presented with intermittent distension and pain in the upper abdomen without other abnormal symptoms. Routine blood examination results were normal. However, abdominal CT revealed a fertile cyst with a diameter of approximately 8 cm, uniform density, and a clear boundary, but without an evident cyst wall in the right lobe of the liver. The cyst was fertile, and phylogenetic analyses revealed that the isolates represented a new E. ortleppi genus haplotype. A result of 10‒14 years incubation period with indigenous infection was considered available for the case through the epidemiological survey.
Conclusions
CE due to E. ortleppi infection can be confused with other diseases causing liver cysts, resulting in misdiagnosis. A transmission chain of E. ortleppi may exist or existed in the past in the previously considered non-endemic areas of echinococcosis in southwestern China.
Graphic abstract

Similar content being viewed by others
Background
Echinococcosis or hydatidosis is a global, often fatal zoonotic helminthic disease caused by Echinococcus sp. (Cestoda: Taeniidae) tapeworm larvae [1]. Intermediate hosts (such as wild herbivores, livestock, and small mammals), as well as accidentally infected humans receive their infections through ingestion of contaminated food and water contaminated by Echinococcus eggs from the feces of the definitive hosts (such as dogs and foxes, etc.) [2]. The eggs hatch to release oncospheres in the intestinal tract, which migrate via the blood circulation to other organs of the body, mainly the liver and lungs, where they develop into hydatid cysts [1, 2]. Presently, nine species of Echinococcus have been reported globally, with six confirmed zoonoses caused by E. granulosus, E. multilocularis, E. vogeli, E. oligarthra, E. canadensis and E. ortleppi; while there is to date no zoonotic evidence for E. equinus, E. felidis and E. shiquicus, which have only been reported to complete their life cycles in different mammals [3, 4].
The main species infecting humans is Echinococcus granulosus, which is globally distributed and generally circulates between dogs and livestock causing cystic echinococcosis (CE) [5]. Echinococcosis remains a significant health problem with a considerable socioeconomic burden in endemic areas, especially in China [5, 6]. Since 1990, several studies focusing on host species, molecular biology, and morphology have posited that E. granulosus is a genotype complex—E. granulosus sensu lato (s.l.), comprising genotypes G1‒G10, and the so-called lion strain. To date, E. granulosus s.l. has been split into five separate species, E. granulosus sensu stricto (s.s.) (G1/G2/G3 genotypes), E. equinus (G4 genotype), E. ortleppi (G5 genotype), E. canadensis group (G6/G7/G8/G10 genotypes), and E. felidis (lion strain) [7,8,9,10,11]. In China, G1 is the predominant genotype, with G3 of E. granulosus s.s. and G6, G7, G8, and G10 of the E. canadensis group recently reported sporadically [12,13,14,15,16].
Echinococcus ortleppi infection was first reported in a herd of cattle in Holland; hence, it was named a cattle strain [7, 8]. To date, reported hosts of E. ortleppi include cattle, camels, pigs, goats, sheep, wild mammals (including oryx, douc langur, crested porcupines and spotted deer) and humans in many regions of Europe, Africa, the Middle East, South Asia, and Latin America [17]. Shi et al. [17] reported in 2019 the first case of a human E. ortleppi infection in Rongshui County, Guangxi Zhuang Autonomous Region, a region of southwestern China previously considered non-endemic. This particular patient had no travel or work history related to any clearly identified epidemic areas. Here, we report a case of E. ortleppi infection in a similar area located in Guizhou Province, bordering the Guangxi is reported. As this constitutes a highly unusual phenomenon, we studied the pathology, phylogeny, and epidemiology of this E. ortleppi infection case.
Methods
Clinical examination and morphological observation
Herein, we reported the case of a 19-year-old female patient who resides in a village of Anshun City, Guizhou, China. She presented first in May 2019 with intermittent distention and pain in the upper abdomen for more than 10 days without evident inducements, such as fever, cough, and vomiting or diarrhoea. On 4 July, she visited the People's Hospital of Anshun City, and routine blood examination and computed tomography (CT) were performed for pathological diagnosis. A surgical treatment plan was implemented, and the cyst including wall and its entire contents of the entire lesion was removed during the implementation. The cystic lesion was harvested after extirpation and fixed it in a 10% formalin solution, subsequently produced paraffin-embedded sections that were stained with hematoxylin–eosin (HE) and observed under the microscope at 40 × magnification for pathological diagnosis.
Molecular identification
The total DNA of the paraffin-embedded specimens was extracted using the QIAamp® DNA FFPE Tissue Kit (56,404, Qiagen, Hilden, Germany) according to the instructions of the manufacturer. We then amplified two mitochondrial DNA (mtDNA) genes; part of cytochrome c oxidase subunit 1 (cox1) and nicotinamide dehydrogenase subunit 1 (nad1). The sequences of the primers used were F/COI (5′-TTG AAT TTG CCA CGT TTG AAT GC-3′) and R/COI (5′-GAA CCT AAC GAC ATA ACA TAA TGA-3′) [18], JB11 (5′- AGA TTC GTA AGG GGC CTA ATA-3′) and JB12 (5′-ACC ACT AAC TAA TTC ACT TTC-3′) [8], respectively. We prepared a 50 μl polymerase chain reaction (PCR) reaction volume consisting of 25 μl of premix Taq polymerase (RR902A, Takara, Dalian, China), 20 μl of RNase-free water (9012, Takara, Dalian, China), 1.5 μl of each primer (10 pmol/μl) (Sangon Biotech Co. Ltd., Shanghai, China), and 2 μl of extracted DNA. Two PCR reactions were performed: 94 °C for 5 min, followed by 35 cycles (denaturation at 94 °C for 30 s, annealing at 52 °C for 45 s for cox1 and 50 °C for 30 s for nad1, extension at 72 °C for 1 min), and an extension step at 72 °C for 10 min, finally saved at 4 °C. The PCR products were cloned and sequenced at BGI-Write Co., Ltd. (Shanghai, China). The resulting sequences were BLAST searched in the NCBI database to identify the species.
Phylogenetic analyses
A total of 49 sequences (27 for the cox1 gene, and 22 for the nad1 gene), including all longer length E. ortleppi sequences obtained in this study and those retrieved from the GenBank (other strains or species of Echinococcus spp.) were used for phylogenetic analysis. The sequences were edited using the Clustal X2 [19] and MAGE 7 [20] software. The substitution saturation of the sequence matrix was tested using DAMBE 5 [21]. We used jModelTest v.2.1.4 [22] to select the best-fit models for nucleotide substitution. Finally, we used the mitochondrial gene of Taenia solium as outgroups to construct Bayesian inference trees of the 875-bp cox1 and 520-bp nad1 genes using MrBayes 3.2.4 [23]. A haplotype network diagram was drawn using Network 5.0 (http://www.fluxus-engineering.com) to analyze the phylogenetic relationship between E. ortleppi isolated in this study and those reported worldwide.
Epidemiological investigation
A detailed survey of the occupation, residence change, travel history, and dog contact experience of the patient, as well as the livestock trade and slaughtering situation around her residence, were conducted through field visits and communication with the patient and her families in November 2019 to determine the sources and time of infection. Additionally, we collected six samples of dog feces from around the residence of the patient to test for Echinococcus via a copro ELISA test using a detection kit for Echinococcus antigen in dog feces (Combined Biotech Co., Ltd., Shenzhen, China).
Results
Pathological characteristics
General medical examination including body temperature (36.9 ºC), pulse (80 /min), blood pressure (130/70 mmHg, 1 mmHg = 0.133 kPa), and respiratory rate (20 breaths/min) of the patient showed values within normal limits. Routine blood examination showed a neutrophil count (6.64 × 109 / L), which was slightly higher than the normal range (1.8‒6.3 × 109 /L), a lymphocyte ratio (17.6%) that was lower than the normal range (20‒50%), the eosinophil count (0.32 × 109 /L) and other measurements were normal (Table 1).
Abdominal CT images revealed that the right lobe of the liver had a spherical low-density shadow with a diameter of approximately 8 cm, the density was uniform, the boundary was clear and the cyst wall was not visible. According to the Manual by the World Health Organization (WHO) and the World Organization for Animal Health (OIE) on Echinococcosis in Humans and Animals [24], the CE classification of the finding was that the cyst was of the CL type with a medium size (5‒10 cm diameter). The HE staining revealed Echinococcus protoscoleces in the cyst contents that were confirmed to be fertile (Fig. 1).
Molecular identification
We sequenced an 875-bp nucleotide sequence numbered as ‘8D21’ for the cox1 gene and a 520-bp sequence numbered as ‘JD11’ for the nad1 gene, which showed 99.66% and 99.81% identify to the two referential E. ortleppi sequences (accession numbers: MK165232 and MN058592, respectively) using BLAST on NCBI. Therefore, it was identified that the isolate in this study was E. ortleppi species. We submitted this information under accession numbers MZ190835 and MZ190836 (the sequences data would be released on December 21, 2021), respectively, to GenBank, the annotated collection of all publicly available DNA sequences at the National Institutes of Health (NIH) in the USA.
Phylogenetic relationships
The Bayesian phylogenetic trees of Echinococcus spp. based on cox1 and nad1 genes showed that the isolates in this study belonged to E. ortleppi, which is separated from other strains of E. granulosus s.l. and other species of Echinococcus. Parsimony network diagrams of E. ortleppi cox1 and nad1 genes revealed the variation between the haplotypes isolated in China and the other global haplotypes. Moreover, the haplotypes were shown to be different between the E. ortleppi reported in the two provinces of Guizhou, and Guangxi, China (Fig. 2).
The Bayesian phylogenetic trees and haplotype networks based on mtDNA genes cox1 a and nad1 (b). The ‘GRT + G + I’ and ‘HKY + G’ substitution models with a 1000-generation sampling interval from 10 000 000 generations of Markov Chain Monte Carlo (MCMC) were set to use Bayesian inferring, respectively. The size of the circle in the network represents the number of sequences of the E. ortleppi haplotype, and the distance between the centers of the circles shows the variation between two haplotypes. The circle color in the network diagrams corresponds to the color of sequence names on the trees
Epidemiologic history
The patient was born in July 2000 in a village of Anshun City, Guizhou Province, and lived there up to the age of nine years. After that, from September 2009, she moved to Ningbo City (Zhejiang Province) and lived until August 2012. She then moved to Pucheng County (Nanping City, Fujian Province), where she lived for three years. During this time, she made a short trip to Shanghai City in 2014. In September 2015, she traveled back to her birthplace, and lived there until the time of the survey. Based on the results of our investigation, she never visited any area identified as endemic for echinococcosis, and she had no contact with dogs from when she lived away from her birthplace.
Additionally, a cattle slaughterhouse was operating from 2005 to 2013 in the village where she was born, and the cattle were mainly brought from the local areas and Guangxi, meanwhile, dogs were always kept in her home. However, in 2019, all six samples of dog faeces collected from around the residence of the patient were negative based on an ELISA detection kit for Echinococcus coproantigens. Therefore, we speculate that there a possible chain of transmission of Echinococcus may exist locally before 2013, which consists of the essential intermediate hosts (the cattle from the slaughterhouse) and the definitive hosts (the dogs from her home). If so, the patient could have been infected with E. ortleppi during this period (from 2004 to 2009), particularly as the latent period of this CE disease was suggested to be 10‒14 years.
A more detail spatiotemporal overview of the patient's life history is shown in an additional figure (see Additional file 1).
Discussion
The patient presented with only abdominal pain without any other symptoms. The most important indicators of the parasitic infection, i.e. count and percentage of eosinophil granulocytes were normal; the cysts had no typical CE (CE1‒CE5) features seen in the CT imagery [24], such as an evident cyst wall structure consistent with the results of a previous study [6, 17]. These preoperative clinical features did not reveal whether the liver cyst was caused by Echinococcus since it was very similar to a congenital liver cyst. Indeed, Maravilla et al. [27] similarly reported a misdiagnosed case of E. ortleppi infection in Mexico. Therefore, differential diagnoses require further diagnostic approaches. The formulation of the surgical plan was affected since protoscoleces could result in severe secondary infections if the cyst turned out to be parasitic in origin and run the risk of rupture during the operation. In the present study, the cyst could be removed without breaking and the subsequent microscopy showed protoscoleces proving it was a fertile Echinococcus cyst.
Molecular diagnosis results revealed that E. ortleppi caused the infection. We speculate that the type CL cyst presentation was either abnormal or that E. ortleppi infection tended to manifest in humans with type CL characteristics. This requires a large sample size to elucidate, which is available in this case. This is the second report on CE caused by E. ortleppi infection in China. In accordance with the first case, it occurred in an area considered to be free of echinococcosis [17]. It has been documented that the main epidemic areas of echinococcosis in the pastoral and semi-pastoral regions of the western China show a large number of Echinococcus genotype G1, with sporadic G3 and G6 type presence but without E. ortleppi (G5) being reported [12,13,14]. The mitochondrial gene sequences of E. ortleppi obtained from the two cases in China are new haplotypes differing from those reported in other regions in the world. This led to the suspicion that the cases were due to local infections, and that there may be, or has been, a chain of transmission of E. ortleppi in the areas of south-western China. This calls for an investigation into the epidemiology and molecular phylogeny of echinococcosis in the southwestern China regions including Guangxi, and Guizhou provinces to search for a potential life cycle of E. ortleppi.
The distribution of E. multilocularis is obviously regional. It is commonly found in high latitude regions of the northern hemisphere (such as Canada, Central Europe, Russian Federation, Mongolia, Hokkaido of Japan and Northern China), alpine regions (such as the Qinghai-Tibet Plateau of China) and permafrost regions (such as Alaska of USA) [28]. However, there is an obvious difference between E. ortleppi and E. multilocularis,according to the statistical results of Shi et al. [17], E. ortleppi infection cases in humans or other intermediate hosts have been reported in Asia, Africa, America and Europe, but, the countries with more than 10 cases reported are mainly in the low-latitude tropical area, such as Brazil (536 cases) in South America [29,30,31,32,33], Ethiopia (13 cases) [29, 34], Kenya (91 cases) [29, 35, 36, Sudan (17 cases) [29, 37], Zambia (53 cases) [29] and Namibia (38 cases) [29] in Africa, and India (12 cases) [2, 38, 39] in South Asia. So, it was inferred that there may be a stable tropical strain of Echinococcus in the long-term environmental adaptive evolution. For China, given the lack of genetic data, it remains elusive whether the E. ortleppi in China originated from the cattle trade across national borders (tropics) in the ancient era or emerged through natural a local selection processes operating over a long time.
Considering that both Guizhou and Guangxi are southern province or region of China, which are close to tropical areas in geographical locations. The warm climate conditions may be favorable for E. ortleppi survival and reproduction, so, here may be a risk of echinococcosis infection. Therefore, we strongly believe that more attention should be paid to echinococcosis in the previously considered non-endemic areas in China and the improve dissemination of health education to the people in the potentially endemic areas. Moreover, the quarantine and management of animals should be enhanced, especially in livestock slaughterhouses, to prevent emerging infections.
However, this investigation only focused on one case, and discussed the transmission chain of E. ortleppi in southwest China in combination with another case previously reported [17]. The sample size was very limited, which also led to the limitations of the inference in this study. According to a report by Han et al. [40], at of the end of 2017, a total of 15 cases and 6 cases were reported in Guizhou Province and Guangxi Zhuang Autonomous Region, respectively. Detailed reports about the infection sources and routes of these cases are still scarce, which restricts the understanding of distribution and transmission of E. ortleppi in China.
Conclusions
In this study, a fertile cyst was formed on the patient’s liver, and it was confirmed as the CE with CL type caused by E. ortleppi. Differentiated inheritance variation of parasite and blank history to stay in the epidemic area identified of patient showed that it might be a locally infected case. To date, the only two E. ortleppi infections found in southwestern China represent a potential transmission pattern, which requires detailed field investigations and laboratory experiments to understand. Moreover, chronic parasitic infectious diseases of this kind pose a high health risk in humans since its atypical pathological features might result in misdiagnosis and spill into other organs during surgery. The infection features, life cycle, and transmission mode of E. ortleppi may be different from those of other Echinococcus spp.; therefore, more studies on the pathology, molecular biology, and epidemiology should be conducted.
Availability of data and materials
The data used in this study are available, if necessary, please contact first author (XW) or corresponding author (WPW).
Abbreviations
- CE:
-
Cystic echinococcosis
- s.l.:
-
Sensu lato
- s.s.:
-
Sensu stricto
- CT:
-
Computed Tomography
- cox1:
-
Cytochrome c oxidase subunit 1
- nad1:
-
Nicotinamide dehydrogenase subunit 1
References
Wang X, Liu J, Zuo Q, Mu Z, Weng X, Sun X, et al. Echinococcus multilocularis and Echinococcus shiquicus in a small mammal community on the eastern Tibetan Plateau: host species composition, molecular prevalence, and epidemiological implications. Parasit Vectors. 2018;11(1):302. https://doi.org/10.1186/s13071-018-2873-x.
Pednekar RP, Gatne ML, Thompson RC, Traub RJ. Molecular and morphological characterisation of Echinococcus from food producing animals in India. Vet Parasitol. 2009;165(1–2):58–65.
Kumaratilake LM, Thompson RCA. A review of the taxonomy and speciation of the genus Echinococcus Rudolphi 1801. Z Parasitenkd. 1982;68(2):121–46.
Xiao N, Qiu J, Nakao M, Li T, Yang W, Chen X, et al. Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China. Int J Parasitol. 2005;35(6):693–701.
Wang Z, Wang X, Liu X. Echinococcosis in China, a review of the epidemiology of Echinococcus spp. EcoHealth. 2008;5(2):115–26.
Craig PS, McManus DP, Lightowlers MW, Chabalgoity JA, Garcia HH, Gavidia CM, et al. Prevention and control of cystic echinococcosis. Lancet Infect Dis. 2007;7(6):385–94.
Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54(2):165–73.
Bowles J, McManus DP. NADH dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus. Int J Parasitol. 1993;23(7):969–72.
Romig T, Ebi D, Wassermann M. Taxonomy and molecular epidemiology of Echinococcus granulosus sensu lato. Vet Parasitol. 2015;213(3–4):76–84.
Kinkar L, Laurimäe T, Sharbatkhori M, Mirhendi H, Kia EB, Ponce-Gordo F, et al. New mitogenome and nuclear evidence on the phylogeny and taxonomy of the highly zoonotic tapeworm Echinococcus granulosus sensu stricto. Infect Genet Evol. 2017;52:52–8.
Hüttner M, Nakao M, Wassermann T, Siefert L, Boomker JD, Dinkel A, et al. Genetic characterization and phylogenetic position of Echinococcus felidis (Cestoda: Taeniidae) from the African lion. Int J Parasitol. 2008;38(7):861–8.
Yang YR, Rosenzvit MC, Zhang LH, Zhang JZ, McManus DP. Molecular study of Echinococcus in West-Central China. Parasitology. 2005;131(Pt 4):547–55.
Zhang LH, Chai JJ, Jiao W, Osman Y, McManus DP. Mitochondrial genomic markers confirm the presence of the camel strain (G6 genotype) of Echinococcus granulosus in north-western China. Parasitology. 1998;116(Pt 1):29–33.
Yan N, Nie HM, Jiang ZR, Yang AG, Deng SJ, Guo L, et al. Genetic variability of Echinococcus granulosus from the Tibetan plateau inferred by mitochondrial DNA sequences. Vet Parasitol. 2013;196(1–2):179–83.
Zhang T, Yang D, Zeng Z, Zhao W, Liu A, Piao D, et al. Genetic characterization of human-derived hydatid cysts of Echinococcus granulosus sensu lato in Heilongjiang Province and the first report of G7 genotype of E. canadensis in humans in China. PLoS ONE. 2014;9(10):e109059. https://doi.org/10.1371/journal.pone.0109059.
Yang D, Zhang T, Zeng Z, Zhao W, Zhang W, Liu A. The first report of human-derived G10 genotype of Echinococcus canadensis in China and possible sources and routes of transmission. Parasitol Int. 2015;64(5):330–3.
Shi Y, Wan X, Wang Z, Li J, Jiang Z, Yang Y. Correction to: First description of Echinococcus ortleppi infection in China. Parasit Vectors. 2019;12(1):412. https://doi.org/10.1186/s13071-019-3677-3.
Nakao M, Sako Y, Yokoyama N, Fukunaga M, Ito A. Mitochondrial genetic code in cestodes. Mol Biochem Parasitol. 2000;111(2):415–24.
Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Xia X. DAMBE5: A Comprehensive Software Package for Data Analysis in Molecular Biology and Evolution. Mol Biol Evol. 2013;30(7):1720–8.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. https://doi.org/10.1038/nmeth.2109.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4.
Eckert J, Gemmell MA, Meslin F-X, Pawlowski ZS. WHO/OIE manual on echinococcosis in humans and animals: a public health problem of global concern. Paris: World Organisation for Animal Health; 2001. p. 32–4.
Zheng CJ, Yang L, Zhang GJ, Wang Q, Wu WW, Yan J. Interpretation of technical scheme for echinococcosis control (Edition 2019). J Trop Dis Parasitol. 2020;18(4):193–201.(in Chinese)
Wu WP, Wang H, Wang Q, Zhou XN, Wang LY, Zheng CJ, et al. A nationwide sampling survey on echinococcosis in China during 2012–2016. Chin J Parasitol Parasit Dis. 2018;36(1):1–14.(in Chinese)
Maravilla P, Andrew Thompson RC, Palacios-Ruiz JA, Estcourt A, Ramirez-Solis E, Mondragon-de-la-Peña C, et al. Echinococcus granulosus cattle strain identification in an autochthonous case of cystic echinococcosis in central Mexico. Acta Trop. 2004;92(3):231–6.
Pawłowski ZS, Eckert J, Vuitton DA, Ammann RW, Kern P, Craig PS, et al. Echinococcosis in humans: clinical aspects, diagnosis and treatment. In: Eckert J, Gemmell MA, Meslin F-X, Pawlowski ZS, editors., et al., WHO/OIE manual on echinococcosis in humans and animals: a public health problem of global concern. Paris: World Organisation for Animal Health; 2001. p. 28–36.
Addy F, Wassermann M, Banda F, Mbaya H, Aschenborn J, Aschenborn O, et al. Genetic polymorphism and population structure of Echinococcus ortleppi. Parasitology. 2017;144(4):450–8.
Balbinotti H, Santos GB, Badaraco J, Arend AC, Graichen DA, Haag KL, et al. Echinococcus ortleppi (G5) and Echinococcus granulosus sensu stricto (G1) loads in cattle from southern Brazil. Vet Parasitol. 2012;188(3–4):255–60.
Monteiro DU, de Azevedo MI, Weiblen C, Ribeiro TC, Emmanouilidis J, Tonin AA, et al. Echinococcus granulosus sensu stricto, Echinococcus canadensis (G7), and Echinococcus ortleppi in fertile hydatid cysts isolated from cattle in Southern Brazil. Acta Trop. 2016;164:41–4.
Hodzic A, Alic A, Supic J, Skapur V, Duscher GG. Echinococcus ortleppi, the cattle strain in a crested porcupine (Hystrix cristata): a new host record. Vet Parasitol. 2018;256:32–4.
de la Rue ML, Takano K, Brochado JF, Costa CV, Soares AG, Yamano K, et al. Infection of humans and animals with Echinococcus granulosus (G1 and G3 strains) and E. ortleppi in southern Brazil. Vet Parasitol. 2011;177(1–2):97–103.
Tigre W, Deresa B, Haile A, Gabriel S, Victor B, Pelt JV, et al. Molecular characterization of Echinococcus granulosus s.l. cysts from cattle, camels, goats and pigs in Ethiopia. Vet Parasitol. 2016;215:17–21.
Dinkel A, Njoroge EM, Zimmermann A, Walz M, Zeyhle E, Elmahdi IE, et al. A PCR system for detection of species and genotypes of the Echinococcus granulosus-complex, with reference to the epidemiological situation in eastern Africa. Int J Parasitol. 2004;34(5):645–53.
Mbaya H, Magambo J, Njenga S, Zeyhle E, Mbae C, Mulinge E, et al. Echinococcus spp. in Central Kenya: a different story. Parasitol Res. 2014;113(10):3789–94.
Ahmed ME, Eltom KH, Musa NO, Ali IA, Elamin FM, Grobusch MP, et al. First report on circulation of Echinococcus ortleppi in the one humped camel (Camelus dromedaries), Sudan. BMC Vet Res. 2013;9:127. https://doi.org/10.1186/1746-6148-9-127.
Sharma M, Sehgal R, Fomda BA, Malhotra A, Malla N. Molecular char- acterization of Echinococcus granulosus cysts in north Indian patients: identification of G1, G3, G5 and G6 genotypes. PLoS Negl Trop Dis. 2013;7: e2262. https://doi.org/10.1371/journal.pntd.0002262.
Chamuah JK, Raina OK, Lalrinkima H, Jacob SS, Sankar M, Sakhrie A, et al. Molecular characterization of veterinary important trematode and cestode species in the mithun Bos frontalis from north-east India. J Helminthol. 2016;90(5):577–82.
Han S, Wu WP, Xue CZ. Analysis of echinococcosis cases from non-endemic areas of China in 2017. J Pathog Biol. 2018;14(8):901–4909. (in Chinese)
Acknowledgements
We thank Wang Ying and Xue Chuizhao (National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention) for their help with data analysis. We are grateful to reviewers for their invaluable comments to improve this manuscript.
Funding
This work was approved by the fund of control and prevention for echinococcosis (2019), and Demonstration project of echinococcosis control (2021) of National Institute of Parasitic Diseases, Chinese Center for Diseases Control and Prevention (Chinese Center for Tropical Diseases Research).
Author information
Authors and Affiliations
Contributions
XW: Performed the molecular experiments and wrote the manuscript. YL, AYZ, JZ, RJ did the epidemiological investigation. HYC, GX, NZX performed clinical examination and morphological observation. BXL: collected and detected the dog feces from around the residence of the patient. WPW, YYG: contributed in the writing of the manuscript. XW, WPW: developing the research scheme. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Ethical Review Committee of National Institute of Parasitic Diseases, Chinese Center for Disease (Chinese Center for Tropical Disease Research) approved this study (identification number: 2021012).
Consent for publication
All presentations of the data have consent for publication.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1.
The traced location and time of the patient. a The roadmap for relocation and travel. b Life history timeline. Statistics of endemic areas originate from the Technical Scheme for Control of Echinococcosis (Edition 2019) [25] and a nationwide sampling survey [26]. Rongshui County is the place of the first description of E. ortleppi infection in China. Anshun City is the birthplace of the patient, and the settlement she lived before September 2009 and after September 2015. Ningbo City is the second settlement (from September 2009 to August 2012), Pucheng County is the third settlement (from August 2012 to September 2015), and Shanghai City is the destination of a short tour in 2014.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Zhu, A., Cai, H. et al. The pathology, phylogeny, and epidemiology of Echinococcus ortleppi (G5 genotype): a new case report of echinococcosis in China. Infect Dis Poverty 10, 130 (2021). https://doi.org/10.1186/s40249-021-00907-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40249-021-00907-3