Abstract
Vaccination of laying hens has been successfully used to reduce egg contamination by Salmonella Enteritidis, decreasing human salmonellosis cases worldwide. Currently used vaccines for layers are either inactivated vaccines or live attenuated strains produced by mutagenesis. Targeted gene deletion mutants hold promise for future vaccines, because specific bacterial functions can be removed that may improve safety and allow differentiation from field strains. In this study, the efficacy of Salmonella Enteritidis ΔtolC and ΔacrABacrEFmdtABC strains in laying hens as live vaccines was evaluated. The mutants are deficient in either the membrane channel TolC (ΔtolC) or the multi-drug efflux systems acrAB, acrEF and mdtABC (ΔacrABacrEFmdtABC). These strains have a decreased ability for gut and tissue colonization and are unable to survive in egg white, the latter preventing transmission of the vaccine strains to humans. Two groups of 30 laying hens were orally inoculated at day 1, 6 weeks and 16 weeks of age with 108 cfu of either vaccine strain, while a third group was left unvaccinated. At 24 weeks of age, the birds were intravenously challenged with 5 × 107 cfu Salmonella Enteritidis PT4 S1400/94. The vaccine strains were not shed or detected in the gut, internal organs or eggs, 2 weeks after the third vaccination. The strains significantly protected against gut and internal organ colonization, and completely prevented egg contamination by Salmonella Enteritidis under the conditions of this study. This indicates that Salmonella Enteritidis ΔtolC and ΔacrABacrEFmdtABC strains might be valuable strains for vaccination of layers against Salmonella Enteritidis.
Similar content being viewed by others
Introduction
Salmonella Enteritidis first emerged in the 1980s as a significant threat to public health worldwide. Eggs were identified as the main food vehicle causing human illness [1, 2]. A sustained commitment of the authorities, implementation of Salmonella control programs and serious investment in Salmonella research led to international progress in decreasing the incidence of both egg contamination [3] and human infections [4]. Vaccination in particular contributed to the decline in the number of recorded human cases of Salmonella Enteritidis [5]. Both inactivated and live vaccines have been shown to reduce Salmonella colonization in layers and contamination of eggs [6–8]. Several live vaccines were developed and proven to be efficient against Salmonella colonization [9–11]. Live vaccines may stimulate both cell-mediated and humoral immunity, can induce rapid protection by colonization-inhibition and are easy to administer, i.e. through the drinking water [6, 12]. A major concern of live vaccines however is safety, including the possible risk of reversion to virulence [13]. Whole gene deletion mutants are generally considered to be less capable of reversion to a virulent phenotype as compared to strains harboring point mutations or undefined genetic alterations. For Salmonella Enteritidis, a lot of knowledge has been generated on the function of many of the chromosomal genes, and targeted deletions of specific genes related to virulence or persistence in a host have been used to construct live vaccine strains [14–18]. In the case of Salmonella vaccines for laying hens, the issue of vaccine safety has an additional dimension, as safety should not only include the target species, but also the risk of transmission to humans through consumption of the eggs. Deleting genes important for virulence in mammals, but also deleting genes that are involved in egg white survival can be a key issue because this will prevent transmission of the vaccine strains to the egg consumers.
Egg white survival is a key characteristic of Salmonella Enteritidis transmission to humans. Because of the low pH, iron restricting conditions and the presence of a variety of antimicrobial molecules, egg white is an antimicrobial matrix [19]. Lipopolysaccharide (LPS) structure [20], lysozyme inhibitors [21] and protein and DNA damage repair mechanisms [22, 23] are important in egg white survival of Salmonella. Deleting genes encoding these functions could thus generate strains with a deficient egg white survival. Recently obtained data suggested that the multi-drug resistance (MDR) pump systems and the TolC outer membrane channel, through which MDR pumps export antibacterial molecules out of the bacterial cell, are also involved in egg white survival [24]. Siderophore export through TolC counteracting iron-deprivation in egg white, or MDR pump-mediated export of antimicrobial molecules out of the bacterial cell may be involved in this [23, 25].
In the current study, we aimed to evaluate the efficiency of the Salmonella Enteritidis ΔtolC and ΔacrABacrEFmdtABC strains, the latter devoid in three MDR efflux pumps, as live vaccines for protection against Salmonella Enteritidis egg contamination and tissue colonization in laying hens.
Materials and methods
Vaccine and challenge strains
The vaccine strains ΔtolC and ΔacrABacrEFmdtABC are defined mutants of Salmonella Enteritidis 147 phage type 4. The wild type strain 147 was originally isolated from egg white and is resistant to streptomycin. The strain is known to colonize the gut and internal organs to a high level [26, 27]. All mutations were constructed according to the one step inactivation method previously described by Datsenko and Wanner [28]. Briefly, for the ΔtolC mutant, a kanamycin resistance cassette, flanked by FRT-sites, was amplified from the pKD4 plasmid with specific primers, homologous with the flanking region of the target gene. The resulting PCR product was used for recombination on the Salmonella Enteritidis 147 strain chromosome using the pKD20 helper plasmid encoding the λ Red system, promoting recombination between the native gene and PCR adjusted antibiotic resistance cassette. Recombinant clones were selected on kanamycin containing plates. Replacement of the target gene by the resistance cassette was confirmed by PCR. The deletion was P22-transduced into a new Salmonella Enteritidis 147 strain. The antibiotic resistance cassette was eliminated using the pCP20 helper plasmid, encoding the FLP-recombinase, mediating recombination between the FRT-sites flanking the kanamycin resistance cassette. For the ΔacrABacrEFmdtABC strain, the procedure was carried out in three steps, successively deleting the acrAB, acrEF and mdtABC genes. P22 transduction was done in the stepwise generated mutants. All targeted genes were completely deleted from start to stop codon, as confirmed by sequencing analysis. Salmonella Enteritidis S1400/94 was used as a challenge strain. The characteristics of this strain have been described previously [29].
The challenge and vaccine strains were incubated overnight with gentle agitation at 37 °C in Luria–Bertani (LB) medium (Sigma, ST. Louis, MO, USA). To determine bacterial titers, tenfold dilutions were plated on brilliant green agar (BGA, Oxford, Basingstoke, Hampshire, UK) for the challenge strain. The vaccine strains were plated on LB supplemented with 1% lactose, 1% phenol red and 100 µg/mL streptomycin to determine the titer, because these strains do not grow on traditional Salmonella culture media. The vaccine and challenge strains were diluted in HBSS (Hanks Balanced Salt Solution, Invitrogen, Paisley, UK) to 108 cfu/mL.
Experimental birds
Ninety (90) day-old Lohmann Brown laying hens (De Biest, Kruishoutem, Belgium) were randomly divided into three groups and housed in separate units. Commercial feed and drinking water was provided ad libitum. The animal experiment in this study followed the institutional guidelines for the care and use of laboratory animals and was approved by the Ethical Committee of the Faculty of Veterinary Medicine, Ghent University, Belgium (EC2013/135). Euthanasia was performed with an overdose of sodium pentobarbital in the wing vein.
Experimental setup
Two different groups (n = 30) of animals were orally immunized at day of hatch, at 6 weeks of age and at 16 weeks of age through crop instillation of 0.5 mL containing 108 cfu Salmonella Enteritidis 147 ΔtolC (group 1) or Salmonella Enteritidis 147 ΔacrABacrEFmdtABC (group 2). A third group of birds (n = 30) was kept as non-immunized but Salmonella challenged positive controls (group 3). At the age of 18 weeks, serum samples were taken for quantification of anti-Salmonella Enteritidis antibodies in an LPS-ELISA [30]. At the same time, cloacal swabs were taken in each group and bacteriologically analyzed for the presence of the vaccine strains. At 21 weeks of age, all the hens were in lay. Eggs were collected daily during 3 weeks for bacteriological detection of the vaccine strain in the egg content. At 24 weeks of age, all the animals were intravenously inoculated in the wing vein with 0.5 mL containing 5 × 107 cfu of the Salmonella Enteritidis challenge strain S1400/94. This protocol was already used previously to produce high levels of internal egg contamination [10, 31]. The eggs were collected daily during 3 weeks after inoculation and analyzed for the presence of the challenge strain. Three weeks after challenge inoculation, all the animals were euthanized by an overdose of pentobarbital in the wing vein. Samples of the spleen, oviduct, ovary, uterus and caecum were aseptically removed for bacteriological quantification of challenge and vaccine strain bacteria.
ELISA to quantify anti-LPS antibodies
For analysis of anti-Salmonella LPS antibodies in serum samples, a previously described indirect ELISA protocol was used [30]. Three 96 well-plates (Sigma) were coated with 100 µL of an LPS solution (10 µg/mL) in 0.05 M carbonate-bicarbonate (pH 9.6; coating buffer) and incubated for 24 h at 4 °C. The LPS was purified from Salmonella Enteritidis PT4 strain. The plates were rinsed four times with phosphate buffered saline (PBS, Sigma) supplemented with 0.1% Tween-20 (Sigma; washing buffer) between each step. In the first step, 100 µL PBS (Sigma) supplemented with 1% bovine serum albumin (BSA, Sigma; blocking buffer) was added to the wells for 1 h at 37 °C. The blocking buffer was then removed. Secondly, serum samples of animals from the different groups were diluted in blocking buffer (1:200) and added to the plates (100 µL). As an internal negative control, serum from a Salmonella free chick was used. Serum from a chick that had been infected experimentally with Salmonella Enteritidis PT4, strain 76Sa88, was used as an internal positive control. The plates were incubated on a shaking platform for 2 h at 37 °C. Thirdly, peroxidase-labelled rabbit anti-chick IgG (100 µL, Sigma) was diluted (1:2000) in blocking buffer and added to the wells for 1 h and 30 min while shaking at 37 °C. Finally 50 µL of TMB substrate (Fisher Scientific, Erembodegem, Belgium) was added to the wells. The reaction was blocked with 50 µL of sulfuric acid (0.5 M). The absorbance was measured in an ELISA reader at 450 nm. Every sample was analyzed in duplicate. Data were shown as S/P ratios, thus [OD(sample) − OD(negative control)]/[OD(positive control) − OD(negative control)]. Negative values were considered as zero.
Bacteriological examination of the challenged birds
Cloacal swabs taken at week 18 were incubated overnight at 37 °C in buffered peptone water (BPW, Oxoid, Basingstoke, Hampshire, UK). Afterwards a loopful was plated on LB plates supplemented with 1% lactose, 1% phenol red and 100 µg/mL streptomycin (Sigma) for the detection of the vaccine strains Salmonella Enteritidis 147 ΔtolC and ΔacrABacrEFmdtABC.
Samples of caecum, spleen, ovary, oviduct and uterus were homogenized in BPW (10% weight/volume suspensions) and tenfold dilutions were made in HBSS (Invitrogen). Six droplets of 20 µL of each dilution were plated on BGA (for quantification of the challenge strain) or on LB supplemented with 1% lactose, 1% phenol red and 100 µg/mL streptomycin (for quantification of the vaccines). After overnight incubation at 37 °C, the number of cfu/g tissue was determined by counting the number of bacterial colonies for the appropriate dilution. Samples that tested negative after direct plating for the challenge strain were enriched in tetrathionate brilliant green broth (Oxoid, Basingstoke, UK) by overnight incubation at 37 °C. After incubation, a loopful of the tetrathionate brilliant green broth was plated on BGA.
Egg production and bacteriological examination of eggs
Eggs were collected daily for 6 weeks from week 21 onwards and the egg production was determined. Each day, eggs of six hens per group were pooled in one batch, yielding an egg per batch number that varied between one and six. Upon collection, lugol solution and 95% ethanol were used to decontaminate the surface of the eggshell. After decontamination of the eggshell, the eggs were broken aseptically and the total content of the eggs was pooled and homogenized per batch. A volume of 40 mL of BPW was added for each egg to the pooled egg content and incubated for 48 h at 37 °C. To detect the vaccine strains, a loopful of the BPW broth was plated on LB plates supplemented with 1% lactose, 1% phenol red and 100 µg/mL streptomycin. To detect the challenge strain, a loopful of the BPW broth was plated on BGA. Additionally, further enrichment was done overnight at 37 °C in tetrathionate brilliant green broth and after incubation, a loopful of broth culture was streaked onto BGA.
Statistical analysis
SPSS 22.0 software was used for statistical analysis. Cloacal swabs, batches of eggs and data of cfu Salmonella/gram tissue of the caecum, spleen, ovary, oviduct and uterus after enrichment were categorized as either positive or negative. A binary regression model was used to determine differences between the groups. For all tests, differences with p values below 0.05 were considered to be statistically significant.
Results
Detection of anti-Salmonella LPS antibodies in serum
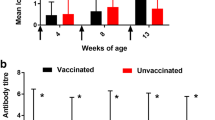
Data derived from the LPS-ELISA are shown in Figure 1. The data are represented as S/P ratios, thus [OD(sample) − OD(negative control)]/[OD(positive control) − OD(negative control)].
[OD(sample) − OD(negative control)]/[OD(positive control) − OD(negative control)] measured in the ELISA detecting anti- Salmonella LPS antibodies. Serum of 18-week old laying hens, vaccinated at day 1, 6 weeks of age and 16 weeks of age with Salmonella Enteritidis 147 ΔtolC and Salmonella Enteritidis 147 ΔacrABacrEFmdtABC was analysed.
Analysis of cloacal swabs and eggs for the presence of vaccine strains
Not a single Salmonella vaccine isolate was obtained from cloacal swabs or egg content samples.
Clinical signs and egg production after challenge
Over the whole experiment, there was no reduction in feed and water intake in either of the groups. The egg production rate after infection in the unvaccinated control group dropped to 59% in the first week post-infection (pi) and raised to 75 and 86% in the second and third week pi. The egg production rate did not decrease significantly after challenge in the vaccinated groups. The egg production percentage in the group vaccinated with the ΔtolC strain was 60, 100 and 90%, in the first, second and third week after challenge. In the group vaccinated with the ΔacrABacrEFmdtABC strain, the egg production percentage was 56, 70 and 68% respectively. Some eggs were thin-shelled and malformed during the first week after infection. At the end of the experiment, 11 hens died in the group of animals vaccinated with the Salmonella Enteritidis 147 ΔacrABacrEFmdtABC strain because of cannibalism.
Isolation of the challenge strain from egg contents
Not a single Salmonella positive egg batch was detected from animals vaccinated with the Salmonella Enteritidis 147 ΔtolC and Salmonella Enteritidis 147 ΔacrABacrEFmdtABC strains (Table 1). During the first week, three egg batches out of 26 were Salmonella positive in the non-vaccinated control group at direct plating. In the third week pi, no positive egg batches were found.
Isolation of the challenge strain from the organs at 3 weeks post-infection
No samples were positive at direct plating. Table 2 presents the percentage of Salmonella-positive organ samples after enrichment, in vaccinated and non-vaccinated groups, at 3 weeks post challenge. Vaccination with the Salmonella Enteritidis 147 ΔtolC strain significantly decreased the number of Salmonella positive samples in the spleen, caecum and ovary as compared to the control group. Vaccination with the ΔacrABacrEFmdtABC strain significantly reduced the number of Salmonella positive samples in the ovary and oviduct.
Discussion
Current commercial live vaccines contain strains harboring undefined mutations in one or more genes on the chromosome or defined point mutations. Strains harboring (undefined or defined) point mutations might, however, revert to a virulent phenotype and are thus considered to be unsafe [13, 32]. Future live vaccines should therefore contain fully defined strains carrying (multiple) gene deletions for purposes of safety. Deletion of entire genes additionally permits differentiation from wild type strains, allowing quality control. Numerous experimental vaccines were already tested in various animal hosts, including chickens, but data on the protection of these live vaccines against egg contamination are scarce [10, 17, 18].
Successful attenuation of the wild type strain requires prior knowledge of the pathogen’s virulence factors. A vaccine strain used for the prevention of (vertical) egg contamination of Salmonella Enteritidis ideally colonizes and induces local immunity in the reproductive tract. From a public health point of view, it may not persist here and preferably does not survive in egg white. A logical approach is to eliminate genes playing a role in egg white survival. In the current study defined mutants in MDR transporters and the TolC outer membrane channel were used as vaccine strains. The TolC promoter is activated after contact with egg white at 42 °C, but not under standard “in vitro” culture conditions [24]. The TolC outer membrane channel is used by MDR transporters (e.g. acrAB, acrEF, mdtABC) to export host antibacterial compounds and bacterial molecules such as siderophores, and is involved in survival in harmful environments, including egg white [33]. The ΔtolC and ΔacrABacrEFmdtABC vaccine strains can no longer survive in egg white, thereby eliminating the risk of human exposure through eggs [24]. To our knowledge, these genes were never associated with protective immunity in chickens, allowing wild type-like antigen presentation.
The actual immune mechanism explaining the protection against Salmonella Enteritidis colonization observed in the current trial is not completely clear. Immunization with Salmonella vaccines can induce variable humoral and cell-mediated responses that do not always correlate with acquired resistance to re-infection [34]. A role for humoral responses in the clearance of Salmonella infections has been shown for using inactivated vaccines, which are less able to induce cellular responses but are still partially protective [35]. Cell-mediated immunity was not investigated during this trial but for Salmonella in poultry, susceptibility to the infection is correlated with a fall in CD4+ and CD8+ T-lymphocytes and γδ T-lymphocytes in the oviduct, and with T-lymphocyte hyporesponsiveness [36]. Live vaccines have been shown to increase numbers of CD4+ and CD8+ T-lymphocytes to a certain level in the gut wall [37]. Future studies should further investigate the role of the humoral and cellular immune responses during vaccine-induced protection. Possibly a combination of cell-mediated immunity and a strong humoral response are yielding additional protective effects.
To conclude, data from this trial indicate that Salmonella Enteritidis ΔtolC and ΔacrABacrEFmdtABC strains are safe vaccines that can induce protection against internal organ colonization after intravenous inoculation of a Salmonella Enteritidis challenge strain. The vaccine strains were able to completely prevent egg contamination with Salmonella Enteritidis in the current in vivo trial.
References
Braden CR (2006) Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin Infect Dis 43:512–517
Greig JD, Ravel A (2009) Analysis of foodborne outbreak data reported internationally for source attribution. Int J Food Microbiol 130:77–87
Esaki H, Shimura K, Yamazaki Y, Eguchi M, Nakamura M (2013) National surveillance of Salmonella Enteritidis in commercial eggs in Japan. Epidemiol Infect 141:941–943
O’Brien SJ (2013) The “decline and fall” of nontyphoidal Salmonella in the United Kingdom. Clin Infect Dis 56:705–710
Cogan TA, Humphrey TJ (2003) The rise and fall of Salmonella Enteritidis in the UK. J Appl Microbiol 94(Suppl):114S–119S
Atterbury RJ, Carrique-Mas JJ, Davies RH, Allen VM (2009) Salmonella colonisation of laying hens following vaccination with killed and live attenuated commercial Salmonella vaccines. Vet Rec 165:493–496
de Freitas Neto OC, Mesquita AL, de Paiva JB, Zotesso F, Berchieri Junior A (2008) Control of Salmonella enterica serovar Enteritidis in laying hens by inactivated Salmonella Enteritidis vaccines. Braz J Microbiol 39:390–396
Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Gast R, Humphrey TJ, Van Immerseel F (2009) Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol Rev 33:718–738
Kilroy S, Raspoet R, Devloo R, Haesebrouck F, Ducatelle R, Van Immerseel F (2015) Oral administration of the Salmonella Typhimurium vaccine strain Nal2/Rif9/Rtt to laying hens at day of hatch reduces shedding and caecal colonization of Salmonella 4,12:i:-, the monophasic variant of Salmonella Typhimurium. Poultry Sci 94:1122–1127
Gantois I, Ducatelle R, Timbermont L, Boyen F, Bohez L, Haesebrouck F, Pasmans F, Van Immerseel F (2006) Oral immunisation of laying hens with the live vaccine strains of TAD Salmonella vac E and TAD Salmonella vac T reduces internal egg contamination with Salmonella Enteritidis. Vaccine 24:6250–6255
Matsuda K, Chaudhari AA, Lee JH (2011) Comparison of the safety and efficacy of a new live Salmonella Gallinarum vaccine candidate, JOL916, with the SG9R vaccine in chickens. Avian Dis 55:407–412
Van Immerseel F, Methner U, Rychlik I, Nagy B, Velge P, Martin G, Foster N, Ducatelle R, Barrow PA (2005) Vaccination and early protection against non-host-specific Salmonella serotypes in poultry: exploitation of innate immunity and microbial activity. Epidemiol Infect 133:959–978
Van Immerseel F, Studholme DJ, Eeckhaut V, Heyndrickx M, Dewulf J, Dewaele I, Van Hoorebeke S, Haesebrouck F, Van Meirhaeghe H, Ducatelle R, Paszkiewicz K, Titball RW (2013) Salmonella Gallinarum field isolates from laying hens are related to the vaccine strain SG9R. Vaccine 31:4940–4945
Parker C, Asokan K, Guard-Petter J (2001) Egg contamination by Salmonella serovar Enteritidis following vaccination with Delta-aroA Salmonella serovar Typhimurium. FEMS Microbiol Lett 195:73–78
De Cort W, Geeraerts S, Balan V, Elroy M, Haesebrouck F, Ducatelle R, Van Immerseel F (2013) A Salmonella Enteritidis hilAssrAfliG deletion mutant is a safe live vaccine strain that confers protection against colonization by Salmonella Enteritidis in broilers. Vaccine 31:5104–5110
De Cort W, Mot D, Haesebrouck F, Ducatelle R, Van Immerseel F (2014) A colonisation-inhibition culture consisting of Salmonella Enteritidis and Typhimurium DeltahilAssrAfliG strains protects against infection by strains of both serotypes in broilers. Vaccine 32:4633–4638
Nassar TJ, Al-Nakhli HM, Al-Ogaily ZH (1994) Use of live and inactivated Salmonella Enteritidis phage type 4 vaccines to immunise laying hens against experimental infection. Rev Sci Tech 13:855–867
Hassan JO, Curtiss R 3rd (1997) Efficacy of a live avirulent Salmonella Typhimurium vaccine in preventing colonization and invasion of laying hens by Salmonella Typhimurium and Salmonella Enteritidis. Avian Dis 41:783–791
Nys Y, Bain M, Van Immerseel F (2011) Improving the safety and quality of eggs and egg products. Woodhead Publishing Limited, Cambridge, pp 183–208
Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F (2009) The Salmonella Enteritidis lipopolysaccharide biosynthesis gene rfbH is required for survival in egg albumen. Zoonoses Public Health 56:145–149
Callewaert L, Aertsen A, Deckers D, Vanoirbeek KG, Vanderkelen L, Van Herreweghe JM, Masschalck B, Nakimbugwe D, Robben J, Michiels CW (2008) A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathog 4:e1000019
Lu S, Killoran PB, Riley LW (2003) Association of Salmonella enterica serovar Enteritidis yafD with resistance to chicken egg albumen. Infect Immun 71:6734–6741
Clavijo RI, Loui C, Andersen GL, Riley LW, Lu S (2006) Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl Environ Microbiol 72:1055–1064
Raspoet R (2014) Survival strategies of Salmonella Enteritidis to cope with antibacterial factors in the chicken oviduct and in egg white. PhD Thesis, Faculty of Veterinary Medicine, Department Pathology, Bacteriology and Poultry Diseases
Li XZ, Plesiat P, Nikaido H (2015) The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418
Methner U, Al-Shabibi S, Meyer H (1995) Infection model for hatching chicks infected with Salmonella Enteritidis. Zentralbl Veterinarmed B 42:471–480
Bohez L, Dewulf J, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F (2008) The effect of oral administration of a homologous hilA mutant strain on the long-term colonization and transmission of Salmonella Enteritidis in broiler chickens. Vaccine 26:372–378
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645
Allen-Vercoe E, Woodward MJ (1999) The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J Med Microbiol 48:771–780
Desmidt M, Ducatelle R, Haesebrouck F, de Groot PA, Verlinden M, Wijffels R, Hinton M, Bale JA, Allen VM (1996) Detection of antibodies to Salmonella Enteritidis in sera and yolks from experimentally and naturally infected chickens. Vet Rec 138:223–226
De Buck J, Van Immerseel F, Haesebrouck F, Ducatelle R (2004) Effect of type 1 fimbriae of Salmonella enterica serotype Enteritidis on bacteraemia and reproductive tract infection in laying hens. Avian Pathol 33:314–320
Audisio MC, Terzolo HR (2002) Virulence analysis of a Salmonella Gallinarum strain by oral inoculation of 20-day-old chickens. Avian Dis 46:186–191
Pan X, Tamilselvam B, Hansen EJ, Daefler S (2010) Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol 10:64
Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G (2001) Salmonella: immune responses and vaccines. Vet J 161:132–164
Feberwee A, de Vries TS, Hartman EG, de Wit JJ, Elbers AR, de Jong WA (2001) Vaccination against Salmonella Enteritidis in Dutch commercial layer flocks with a vaccine based on a live Salmonella Gallinarum 9R strain: evaluation of efficacy, safety, and performance of serologic Salmonella tests. Avian Dis 45:83–91
Johnston CE, Hartley C, Salisbury AM, Wigley P (2012) Immunological changes at point-of-lay increase susceptibility to Salmonella enterica serovar Enteritidis infection in vaccinated chickens. PLoS One 7:e48195
Berndt A, Methner U (2001) Gamma/delta T cell response of chickens after oral administration of attenuated and non-attenuated Salmonella typhimurium strains. Vet Immunol Immunopathol 78:143–161
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SK participated in the design of the study, performed the experiments, analyzed the data and drafted the manuscript. RR, FVI, RD and FH coordinated the study, participated in the design of the study, helped to interpret the results and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank all colleagues that helped during sampling.
Funding
This work was supported by Elanco Animal Health and by the Flemish Agency for Innovation by Science and Technology (IWT), Grant No. 121095.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kilroy, S., Raspoet, R., Haesebrouck, F. et al. Prevention of egg contamination by Salmonella Enteritidis after oral vaccination of laying hens with Salmonella Enteritidis ΔtolC and ΔacrABacrEFmdtABC mutants. Vet Res 47, 82 (2016). https://doi.org/10.1186/s13567-016-0369-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-016-0369-2