Abstract
Background
Campylobacter jejuni is a zoonotic pathogen that infects the human gut through the food chain mainly by consumption of undercooked chicken meat, raw chicken cross-contaminated ready-to-eat food or by raw milk. In the last decades, C. jejuni has increasingly become the most common bacterial cause for food-born infections in high income countries, costing public health systems billions of euros each year. Currently, different whole genome sequencing techniques such as short-read bridge amplification and long-read single molecule real-time sequencing techniques are applied for in-depth analysis of bacterial species, in particular, Illumina MiSeq, PacBio and MinION.
Results
In this study, we analyzed a recently isolated C. jejuni strain from chicken meat by short- and long-read data from Illumina, PacBio and MinION sequencing technologies. For comparability, this strain is used in the German PAC-CAMPY research consortium in several studies, including phenotypic analysis of biofilm formation, natural transformation and in vivo colonization models. The complete assembled genome sequence most likely consists of a chromosome of 1,645,980 bp covering 1665 coding sequences as well as a plasmid sequence with 41,772 bp that encodes for 46 genes. Multilocus sequence typing revealed that the strain belongs to the clonal complex CC-21 (ST-44) which is known to be involved in C. jejuni human infections, including outbreaks. Furthermore, we discovered resistance determinants and a point mutation in the DNA gyrase (gyrA) that render the bacterium resistant against ampicillin, tetracycline and (fluoro-)quinolones.
Conclusion
The comparison of Illumina MiSeq, PacBio and MinION sequencing and analyses with different assembly tools enabled us to reconstruct a complete chromosome as well as a circular plasmid sequence of the C. jejuni strain BfR-CA-14430. Illumina short-read sequencing in combination with either PacBio or MinION can substantially improve the quality of the complete chromosome and epichromosomal elements on the level of mismatches and insertions/deletions, depending on the assembly program used.
Similar content being viewed by others
Background
Campylobacter jejuni is a Gram-negative bacterium that colonizes a wide range of hosts as part of the natural gut microbiota [1]. It is frequently found in farm animals such as chicken and cattle or in wild birds. While consuming undercooked poultry meat, unpasteurized milk or cross-contaminated ready-to-eat food it can colonize the human gut and cause an infectious gastroenteritis together with diarrhea, fever and cramps [2].
Over the past two decades the incidence of Campylobacter infections has continued to increase worldwide and has become a dangerous threat to public health. To date, campylobacteriosis is the most common bacterial cause of food-born infections in high income countries, with costs amounting to 2.4 billion euros each year for the public health system and lost productivity in the European Union [3].
The BfR-CA-14430 strain was first isolated during the zoonosis monitoring program, in which distinct matrix–pathogen combinations were collected by federal state laboratories. The strain was isolated from a German chicken meat sample in August 2016 using ISO 10272-1:2006 [4]. Since this strain was chosen to serve as a fresh field strain for the German research consortium PAC-CAMPY, we analyzed characteristics of BfR-CA-14430, like antibiotic resistance and virulence factors. In addition, we gained a deeper insight into whole genome sequencing and the impact of various assembly programs, including different hybrid assemblers on various combinations of long and short read sequencing technologies. This revealed a complete chromosomal sequence as well as one closed plasmid sequence.
Methods
Bacterial isolation and initial characterization
BfR-CA-14430 was isolated in the framework of the zoonosis monitoring program 2016 from chicken meat according to ISO 10272-1:2006. Species identification was performed by Real-time PCR according to Mayr et al. [5]. The multi locus sequence type was determined by Sanger sequencing (PubMLST) and confirmed by whole-genome sequencing (WGS). The flagellin subunit A (flaA) type was Sanger sequenced [6], typing was done according to PubMLST (pubmlst.org) and compared with the outcome of the WGS analysis. BfR-CA-14430 was cultured either on Columbia blood agar (Oxoid) or in brain heart infusion (Oxoid) at 42 °C under microaerobic conditions (5% O2, 10% CO2) and cells were harvested by centrifugation.
Antimicrobial resistance determination by microdilution
BfR-CA-14430 was pre-cultured on Columbia blood agar for 24 h at 42 °C under microaerobic atmosphere. Broth microdilution susceptibility testing was performed according to VET06 and M45-A [7]. 2–8 × 105 CfU/ml were inoculated into cation-supplemented Mueller Hinton broth (TREK Diagnostic Systems, UK) supplemented with 5% fetal calf serum (PAN-Biotech, Germany), into the European standardized microtiter EUCAMP2 or EUVSEC plate formats (TREK Diagnostic Systems). Samples were incubated for 48 h at 37 °C under microaerobic conditions. Minimal inhibitory concentrations (MIC; [mg/l]) were semi-automatically analyzed using the Sensititre Vizion system and the SWIN-Software (TREK Diagnostic Systems). Epidemiological cut-off values for resistance determination were based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST.org), if already defined for C. jejuni or, alternatively, for Salmonella (EUVSEC plate format).
Genomic DNA extraction and sequencing
DNA extraction for Sanger MLST analyses was performed with GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific). DNA for WGS was prepared using the MagAttract HMW Genomic Extraction Kit (Qiagen) (for PacBio and Illumina sequencing) and QIAamp DNA Mini Kit (Qiagen) for MinION sequencing and further concentrated by precipitation with 0.3 M sodium acetate pH 5 and 0.7 volume isopropanol at room temperature for 30 min. After centrifugation and washing of the precipitate with 70% ice-cold ethanol, the DNA was dissolved in Tris buffer pH 7.5. The quality of the DNA was evaluated by spectral analysis (NanoDrop Spectrophotometer, Thermo Fisher Scientific, USA) and the concentration was fluorimetrically quantified to be 110 ng/µl by Qubit 3.0 Fluorometer (dsDNA BR Assay Kit; Invitrogen, USA). DNA was additionally controlled for lack of sheering products < 20 kb on a 0.8% agarose gel. Sequencing was performed on a MiSeq sequencer (MiSeq Reagent Kit v.3; Illumina Inc., San Diego, CA, USA), using the Library Preparation kit Nextera XT (Illumina Inc., San Diego, CA, USA) resulting in 300-bp paired-end reads and an average coverage of around 100-fold. Furthermore, size selection was performed using 10 K Blue Pippin and DNA was sequenced with Single Molecule Real-Time (SMRT) Sequencing Technology on a PacBio RS II by GATC Biotech AG (Konstanz, Germany) as well as with long read sequencing on Oxford Nanopore MinION (Oxford, UK) (Library-Kit: Rapid Barcoding Kit (SQK-RBK004), Flowcell: 1D R9.4, without size selection, base calling with albacore v2.1.0) in order to compare these three techniques for establishing a complete genome with epichromosomal elements. Total amounts of extracted DNA of 1 ng, 5 µg and 400 ng was used as starting material for sequencing by MiSeq, PacBio or MinION, respectively. A general overview of the raw data from the different sequencing machines can be found in Table 1.
Genome assembly and annotation
Sequencing reads obtained from the MiSeq sequencer were (i) assembled by the SPAdes v3.12 [8] and plasmidSPAdes [9] assembler or (ii) used to correct long read data. Furthermore we used the CLC Genomics Workbench v12.0.1 as well as an assembly from the PacBio in-house pipeline HGAP v3.0 [10] and Flye v2.5 [11] for the PacBio long read assemblies. The assembly based on MinION raw reads was only performed by Flye v2.5. All assemblers were run with default settings. To generate an optimal assembly and derive a closed genome sequence we tested various de novo hybrid assembly tools on different combinations of short and long reads (Unicycler v0.4.7 [12] and wtdbg2 v2.1 [13]). Unicycler first creates a draft genome assembly with SPAdes v3.12 and connects the contigs only afterwards by using the long reads from PacBio or MinION. Wtdbg2, on the other hand, first assembles the long reads and corrects the assembly afterwards by mapping the short reads against the genome. Long reads were mapped to the genomes by minimap2 v2.14 [14]. The different combinations of short and long reads used for each tool are shown in Table 2. In order to annotate the genomes, a custom-made database of 137 complete genomes of C. jejuni downloaded from NCBI (Additional file 1: Table S1) was built and used as a Genus-specific BLAST database for Prokka v1.13 [15].
Assembly comparison and in silico analysis
The assembled genomes were compared by the progressive Mauve algorithm [16] to detect major structural differences. Single nucleotide polymorphisms (SNPs) were detected by mapping the Illumina paired-end reads against the assemblies by bowtie2 v4.8.2 [17] with the end-to-end sensitive mode. SNPs, insertions and deletions were counted within an allele frequency of at least 75% at positions with a minimum of 10 reads by freebayes v.1.2.0 [18] according to Illumina short reads. The multi locus sequence typing (MLST) was performed by a BLAST based pipeline (https://github.com/tseemann/mlst) to identify the allele variants of the seven housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt and uncA). Point mutations conferring antibiotic resistance or individual antibiotic resistance genes were revealed by ResFinder 3.0 [19] (CGE, DTU, Lyngby, DK; https://cge.cbs.dtu.dk/services/ResFinder/).
Quality assurance
In order to perform an in-silico control for contamination within the sequenced DNA, Illumina short reads were adapter trimmed with Flexbar [20] and all reads were taxonomically classified as C. jejuni by Kraken v2.0.6 [21]. Taxonomic classification of the long reads could identify 3.71% of Human related DNA within the PacBio read, which has been removed. Assembly completeness and contamination was controlled with checkM v. 1.0.18 [22].
Results
Antimicrobial resistance profile of BfR-CA-14430
The minimal inhibitory concentration (MIC) of different antibiotics was determined using the broth microdilution susceptibility approach (CLSI). Using the standard EUCAMP2 plate format, which is used for screening of C. jejuni resistance during zoonosis monitoring, the strain showed resistance against ciprofloxacin, nalidixic acid and tetracycline but was sensitive towards erythromycin, gentamicin and streptomycin. We extended the antimicrobial substances and applied the EUVSEC plate format, usually tested with Salmonella and Escherichia coli isolates. As C. jejuni is intrinsically resistant against most of the cephalosporine antibiotics, it was expected that strain BfR-CA-14430 was also resistant against cefotaxime, cefoxitime, cefepime, ceftazidime. The cephalosporine cefoperazone is used as a selective supplement in ISO 10272:2017 in mCCDA (modified charcoal-cefoperazone agar) and Bolton broth. Besides, the strain revealed natural resistance against trimethroprim due to the absence of the target dihydrofolate reductase (FolA). However, MIC values for sulfamethoxazole were 16 mg/l, rendering the strain sensitive, on the basis of a cut-off value used for Salmonella of 64 mg/l. Furthermore, resistance against ampicillin was also seen with MIC values > 64 mg/l, while MIC values for meropeneme, ertapeneme and colistin were 0.25 and 0.5 and 2 mg/l, respectively. BfR-CA-14430 was fully susceptible to chloramphenicol, tigecycline, azithromycin and imipeneme, with MIC values below the lowest test concentration.
Genomic features of the strain BfR-CA-14430
Using multilocus sequence typing, the strain BfR-CA-14430 was identified as sequence type ST-44 which belongs to the clonal complex CC-21 that is frequently found in human infections and well known to cause C. jejuni outbreaks [23]. The complete genome sequence, assembled from MinION and Illumina reads by Unicycler, consists of one chromosome of 1,645,980 bp covering 1,665 coding sequences (CDSs), including blaOXA-61 (Cj0299 in NCTC 11168) that encodes for a beta-lactam resistance gene [24] and a point mutation in the gyrase subunit A (gyrA) (T86I) [25], conferring resistance against (fluoro-)quinolones. All AMR genes or AMR associated SNPs could be detected within the hybrid assembly as well as in the Illumina paired-end reads. Additionally, the genome has 44 transfer RNA (tRNA) genes, 9 ribosomal RNA (rRNA) genes forming three identical operons consisting of 16S, 23S and 5S subunits and an overall GC content of 30.4%. The chromosome harbors the virulence factors cdtA, cdtB, cdtC, coding for the cytolethal distending toxin, the gene encoding the fibronectin-binding protein CadF and the Campylobacter invasion antigens CiaB and CiaC. Genes encoding the monofunctional α 2,3-sialyltransferase CstIII and the N-acetylneuraminic acid biosynthesis proteins NeuA1, NeuB1 and NeuC1 are present for lipooligosacharide (LOS) sialylation, which was shown to be linked to Guillain–Barré syndrome onset [26, 27]. The conserved capsule biosynthesis kpsC and kpsF genes flank the variable capsule locus of approximately 26 kb, belonging to the Penner type HS1 complex [28]. Besides, the pseA-I genes involved in flagellar protein glycosylation [29] were detected on the chromosome. Furthermore, the strain carries a single circular plasmid of 41,772 bp including 46 CDSs. Among these genes the plasmid carries a tetO gene for tetracycline resistance as well as virB2-11 and virD4 genes encoding for a putative type IV secretion system (T4SS), for conjugative DNA transfer between Campylobacter strains [30]. The plasmid showed 93% identity and 98% coverage with plasmid pTet from C. jejuni strain 81–176 (45,025 bp) (CP000549) and 98% identity and 97% coverage with plasmid pMTVDSCj16-1 (42,686 bp) from C. jejuni strain MTVDSCj16 (NZ_CP017033.1) that carry type IV secretion systems and tetO genes as well [31]. By mapping of Illumina paired-end reads, plasmid pMTVDSCj16-1 was covered by 97% with 99% identity and 611 SNPs. Two regions of 600 bp and 1113 bp were not covered by the Illumina reads. However, read mapping was not able to detect a region 927 bp containing a CDS that can also be found in pTet-M129 (NZ_CP007750.1) [32] of C. jejuni strain M129 (NZ_CP007749.1) and pRM5611 (NZ_CP007180.1) from C. coli strain RM5611 (NZ_CP007179.1).
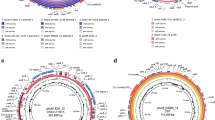
The genomic structure and annotation of the chromosome and plasmid are visualized in Fig. 1 and can be accessed at the National Center for Biotechnology Information (NCBI) database with the accession numbers CP043763 and CP043764.
Genome map, generated by CGView [33], of chromosomal DNA a) and plasmid DNA b) from C. jejuni. BfR-CA-14430. Circles form outside to inside showing: (1,2) coding regions (light blue) predicted on forward (outer circle) and reverse strands (inner circle); (3) tRNAs (dark red); (4) rRNAs (light green); (5) regions above (green) and below (purple) the average GC skew; (6) GC content (black) and (7) DNA coordinates
Assembly comparison
Whole genome comparison of all assemblies showed that each assembler created one chromosome of around 1.6 Mb as well as one plasmid of around 42 kb while using PacBio or MinION long reads in combinations with Illumina short reads (Table 2). Gel electrophoresis of extracted DNA from BfR-CA-14430 suggested the occurrence of chromosomal and plasmid DNA. All long read assembler reconstructed the chromosomal genome in one single contig without large structural variations (Fig. 2). Reads from MinION and Illumina that were assembled by Unicycler resulted in a circular genome. However, some tools generated small extra contigs (Table 2): The combination of Illumina and PacBio data as well as MinION with Illumina data as input to the wtdgb2 assembler generated contigs that were later identified by BLAST to be part of the chromosomal sequence of the strain. With the advantage of using long reads, one misassembly inside a repeat region in the SPAdes assembly based on the Illumina short reads was discovered (Fig. 2). Additionally, we were able to identify the Sanger sequenced flaA gene with a sequence identity of 100% in most of the cases (Table 2). The MinION assembly generated with Flye did not reach 100% sequence identity, due to the high number of SNPs within this assembly.
Progressive Mauve Alignment of chromosomal genomes generated by different assemblers. The Misassembly made by SPAdes is marked by the red square. Assemblies are index by alphabetic letters as shown in Table 2. Color coded blocks indicating homology between the genomes
Furthermore, all tools assembled a plasmid with a size of around 42 kb, except from the PacBio in-house pipeline that created a 64 kb plasmid. By performing a global alignment against itself and generating a dotplot we could show a large repeat region between the first and the last 20 kb in the circular sequence that obviously originates from an assembly error (Fig. 3). Plasmid assemblies produced by Unicycler were found to be circularized, while using PacBio as well as MinION data. Identification of plasmid sequences by plasmidSPAdes, revealed 9 from 3 components. Besides the ca. 42 kb plasmid described earlier, the 8 other sequences could be identified as part of the chromosomal DNA by BLAST from strain BfR-CA-11430 as well as in several closed genomes from Additional file 1: Table S1. Those assembled DNA fragments mainly have their origin in low coverage or repeat regions, which cannot be resolved by short reads and is known to lead to misassemblies in plasmidSPAdes [9].
The dotplot shows a global alignment of the plasmid sequence, generated from PacBio reads by HGAP (Table 2B), against itself. This revealed one dark blue diagonal line in the middle from start to end of the sequence as well as two additional dark blue lines showing up in the top left and bottom right part of the plot. Those lines show a repeat from 42 to 65 kb and 1 to 23 kb, respectively. Therefore, the sequence is identical in the first 23 kb as well as the last 23 kb and indicates it as a large repeat region that is likely to be cause through an assembly error
Standalone assemblies of long read data from MinION generated the overall correct structure of the genome and the plasmid, but a lot of small insertions, deletions and SNPs were additionally created (Table 2). The assembly of MinION raw reads contains more than 25,000 SNPs, which is around 100 times more compared to assemblies of PacBio reads with HGAP and Flye. However, by combining MinION with Illumina data the SNP count decreased to only 20 SNPs. The assembly from HGAP or Flye based on PacBio raw reads contains 155 SNPs and 255 SNPs respectively whereas the combination of PacBio and Illumina contains 0 SNPs.
The final chromosomal assembly of MinION and Illumina reads is covered by 95×, 424× and 375×, whereas the plasmid sequence is covered by 204×, 291× and 3021× from Illumina, PacBio and MinION reads. Genome completeness was calculated to be at 99.36% and contamination was predicted to be 0.15%.
Conclusion
Here, we describe the C. jejuni strain BfR-CA-14430 that carries a beta lactamase and tetracycline resistance gene as well as potential virulence factors that might play a role in human gut infection. Furthermore we compared multiple hybrid assembly methods based on different sequencing technologies. This revealed that the combination of long reads with short reads decreases the SNP rate in de novo assemblies to a large extent. In general, using a combination of long and short reads as input to the Unicycler assembler resulted in accurate and closed chromosomal and plasmidal sequences for our data. However, assemblies based only on PacBio reads, seem to be highly accurate and can also be used without being polished by Illumina data.
Availability of data and materials
The completed genome sequence of BfR-CA-14430 has been deposited into GenBank database with accession number CP043763 (chromosome) and CP043764 (plasmid), respectively. Raw read data from Illumina Miseq, PacBio RS II and Oxford Nanopore MinION is available at NCBI with SRA accession number PRJNA562653.
References
Young KT, Davis LM, DiRita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–79.
Humphrey T, O’Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117(3):237–57.
Food Safety Authority E, Boelaert F, Van der Stede Y, Nagy K, Rizzi V, Garcia Fierro R, et al. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016 Acknowledgements: EFSA and the ECDC wish to thank the members of the Scientific Network for Zoonoses Monitoring Data and the Food and Wat. EFSA J. 2017;15(12):5077.
Jacobs-Reitsma WF, Jongenburger I, de Boer E, Biesta-Peters EG. Validation by interlaboratory trials of EN ISO 10272—microbiology of the food chain—horizontal method for detection and enumeration of Campylobacter spp.—part 2: Colony-count technique. Int J Food Microbiol. 2019;288:32–8.
Mayr AM, Lick S, Bauer J, Thärigen D, Busch U, Huber I. Rapid detection and differentiation of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari in food, using multiplex real-time PCR. J Food Protect. 2010;73(2):241–50.
Wassenaar TM, Newell DG. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9.
Clinical and Laboratory Standards Institute. VET06: methods for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria isolated from animals. 1st ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. {SPAdes}: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32(22):3380–7.
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–9.
Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–6.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595.
Ruan J, Li H. Fast and accurate long-read assembly with wtdbg2. bioRxiv. 2019. https://doi.org/10.1101/530972.
Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9.
Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE ONE. 2010;5(6):e11147.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint. 2012. arXiv:1207.3907.
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4.
Roehr JT, Dieterich C, Reinert K. Flexbar 3.0—{SIMD} and multicore parallelization. Bioinformatics. 2017;33(18):2941–2.
Wood DE, Salzberg SL. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Sails AD, Swaminathan B, Fields PI. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J Clin Microbiol. 2003;41(10):4733–9.
Griggs DJ, Peake L, Johnson MM, Ghori S, Mott A, Piddock LJV. β-lactamase-mediated β-lactam resistance in Campylobacter species: prevalence of Cj0299 (blaOXA-61) and evidence for a novel β-lactamase in C. jejuni. Antimicrob Agents Chemother. 2009;53(8):3357–64.
Luo N, Sahin O, Lin J, Michel LO, Zhang Q. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother. 2003;47(1):390–4.
Guerry P, Ewing CP, Hickey TE, Prendergast MM, Moran AP. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect Immun. 2000;68(12):6656–62.
Godschalk PCR, Heikema AP, Gilbert M, Komagamine T, Wim Ang C, Glerum J, et al. The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barré syndrome. J Clin Investig. 2004;114(11):1659–65.
Poly F, Serichatalergs O, Schulman M, Ju J, Cates CN, Kanipes M, et al. Discrimination of major capsular types of Campylobacter jejuni by multiplex PCR. J Clin Microbiol. 2011;49(5):1750–7.
Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, et al. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60(2):299–311.
Cao TB, Saier J. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: Organismal distribution, structural constraints and evolutionary conclusions. Microbiol Microbiol Soc. 2001;147:3201–14.
Taveirne ME, Dunham DT, Perault A, Beauchamp JM, Huynh S, Parker CT, et al. Complete annotated genome sequences of three Campylobacter jejuni strains isolated from naturally colonized Farm-Raised chickens. Genome Announc. 2017;5(4):e01407-16.
Day WA, Pepper IL, Joens LA. Use of an arbitrarily primed PCR product in the development of a Campylobacter jejuni-specific PCR. Appl Environ Microbiol. 1997;63:1019–23.
Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(Web Server issue):W181–W184184.
Acknowledgements
Not applicable.
Funding
Funding was received by the German Federal Ministry of Education and Research (BMBF) and by the German Federal Ministry of Health (BMG) for the PAC-CAMPY and MinION-Zoo research project and by BfR. L.E. and T.S. were financed in PAC-CAMPY IP10/01KI1725F and MinION-Zoo ZMVI1-2519NIK704 and J.G. and K.S. in PAC-CAMPY IP3/01KI1725B and BfR-SFB-1332-646. The funders had no influence in the study design, data analysis or publication.
Author information
Authors and Affiliations
Contributions
LE, KS, LHW and TS designed the study. JCG, MTK CH and AT performed the laboratory work. LE and TS did the bioinformatics analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Campylobacter jejuni Genomes used to build the Prokka Database
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Epping, L., Golz, J.C., Knüver, MT. et al. Comparison of different technologies for the decipherment of the whole genome sequence of Campylobacter jejuni BfR-CA-14430. Gut Pathog 11, 59 (2019). https://doi.org/10.1186/s13099-019-0340-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-019-0340-7